Predicting virus-host association by Kernelized logistic matrix factorization and similarity network fusion
- PMID: 31787095
- PMCID: PMC6886165
- DOI: 10.1186/s12859-019-3082-0
Predicting virus-host association by Kernelized logistic matrix factorization and similarity network fusion
Abstract
Background: Viruses are closely related to bacteria and human diseases. It is of great significance to predict associations between viruses and hosts for understanding the dynamics and complex functional networks in microbial community. With the rapid development of the metagenomics sequencing, some methods based on sequence similarity and genomic homology have been used to predict associations between viruses and hosts. However, the known virus-host association network was ignored in these methods.
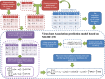
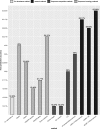
Results: We proposed a kernelized logistic matrix factorization with integrating different information to predict potential virus-host associations on the heterogeneous network (ILMF-VH) which is constructed by connecting a virus network with a host network based on known virus-host associations. The virus network is constructed based on oligonucleotide frequency measurement, and the host network is constructed by integrating oligonucleotide frequency similarity and Gaussian interaction profile kernel similarity through similarity network fusion. The host prediction accuracy of our method is better than other methods. In addition, case studies show that the host of crAssphage predicted by ILMF-VH is consistent with presumed host in previous studies, and another potential host Escherichia coli is also predicted.
Conclusions: The proposed model is an effective computational tool for predicting interactions between viruses and hosts effectively, and it has great potential for discovering novel hosts of viruses.
Keywords: Gaussian interaction profile; Logistic matrix factorization; Oligonucleotide frequency; Similarity network fusion; Virus-host association.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Prediction of Virus-Receptor Interactions Based on Improving Similarities.J Comput Biol. 2021 Jul;28(7):650-659. doi: 10.1089/cmb.2020.0544. Epub 2021 Jan 21. J Comput Biol. 2021. PMID: 33481654
-
Prediction of virus-host infectious association by supervised learning methods.BMC Bioinformatics. 2017 Mar 14;18(Suppl 3):60. doi: 10.1186/s12859-017-1473-7. BMC Bioinformatics. 2017. PMID: 28361670 Free PMC article.
-
Multi-network logistic matrix factorization for metabolite-disease interaction prediction.FEBS Lett. 2020 Jun;594(11):1675-1684. doi: 10.1002/1873-3468.13782. Epub 2020 Apr 26. FEBS Lett. 2020. PMID: 32246474
-
Computational approaches to predict bacteriophage-host relationships.FEMS Microbiol Rev. 2016 Mar;40(2):258-72. doi: 10.1093/femsre/fuv048. Epub 2015 Dec 9. FEMS Microbiol Rev. 2016. PMID: 26657537 Free PMC article. Review.
-
From deep sequencing to viral tagging: recent advances in viral metagenomics.Bioessays. 2013 May;35(5):436-42. doi: 10.1002/bies.201200174. Epub 2013 Mar 1. Bioessays. 2013. PMID: 23450659 Review.
Cited by
-
Improving the reporting of metagenomic virome-scale data.Commun Biol. 2024 Dec 20;7(1):1687. doi: 10.1038/s42003-024-07212-3. Commun Biol. 2024. PMID: 39706917 Free PMC article.
-
Host prediction for disease-associated gastrointestinal cressdnaviruses.Virus Evol. 2022 Sep 16;8(2):veac087. doi: 10.1093/ve/veac087. eCollection 2022. Virus Evol. 2022. PMID: 36325032 Free PMC article.
-
A multitask transfer learning framework for the prediction of virus-human protein-protein interactions.BMC Bioinformatics. 2021 Nov 27;22(1):572. doi: 10.1186/s12859-021-04484-y. BMC Bioinformatics. 2021. PMID: 34837942 Free PMC article.
-
Computational analysis of fused co-expression networks for the identification of candidate cancer gene biomarkers.NPJ Syst Biol Appl. 2021 Mar 12;7(1):17. doi: 10.1038/s41540-021-00175-9. NPJ Syst Biol Appl. 2021. PMID: 33712625 Free PMC article.
-
iPHoP: An integrated machine learning framework to maximize host prediction for metagenome-derived viruses of archaea and bacteria.PLoS Biol. 2023 Apr 21;21(4):e3002083. doi: 10.1371/journal.pbio.3002083. eCollection 2023 Apr. PLoS Biol. 2023. PMID: 37083735 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources