ShORR-1, a Novel Tomato Gene, Confers Enhanced Host Resistance to Oidium neolycopersici
- PMID: 31787994
- PMCID: PMC6854008
- DOI: 10.3389/fpls.2019.01400
ShORR-1, a Novel Tomato Gene, Confers Enhanced Host Resistance to Oidium neolycopersici
Abstract
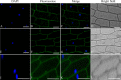
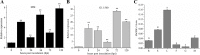
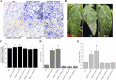
A previous complementary cDNA-amplified fragment length polymorphism (cDNA-AFLP) analysis examined responses to the powdery mildew pathogen Oidium neolycopersici (On) of the resistant cultivar Solanum habrochiates G1.1560, carrying the Ol-1 resistance gene, and susceptible cultivar S. lycopersicum Moneymaker (MM). Among other findings, a differentially expressed transcript-derived fragment (DE-TDF) (M14E72-213) was upregulated in near isogenic line (NIL)-Ol-1, but absent in MM. This DE-TDF showed high homology to a gene of unknown function, which we named ShORR-1 (Solanum habrochaites Oidium Resistance Required-1). However, MM homolog of ShORR-1 (named ShORR-1-M) was still found with 95.26% nucleic acid sequence similarity to ShORR-1 from G1.1560 (named ShORR-1-G); this was because the cut sites of restriction enzymes in the previous complementary cDNA-AFLP analysis was absent in ShORR-1-M and differs at 13 amino acids from ShORR-1-G. Transient expression in onion epidermal cells showed that ShORR-1 is a membrane-localized protein. Virus-induced gene silencing (VIGS) of ShORR-1-G in G1.1560 plants increased susceptibility to On. Furthermore, overexpressing of ShORR-1-G conferred MM with resistance to On, involving extensive hydrogen peroxide accumulation and formation of abnormal haustoria. Knockdown of ShORR-1-M in MM did not affect its susceptibility to On, while overexpressing of ShORR-1-M enhanced MM's susceptibility to On. We also found that changes in transcript levels of six well-known hormone signaling and defense-related genes are involved in ShORR-1-G-mediated resistance to On. The results indicate that ShORR-1-M and ShORR-1-G have antagonistic effects in tomato responses to On, and that ShORR-1 is essential for Ol-1-mediated resistance in tomato.
Keywords: H2O2 accumulation; Oidium neolycopersici; Ol-1-mediated resistance; abnormal haustoria; resistant tomato; susceptible tomato.
Copyright © 2019 Zhang, Xu, Pei, Yu, Zhang, Li, Chen, Yang, Zhou and Li.
Figures







References
-
- Bai Y., Huang C. C., van der Hulst R., Meijer-Dekens F., Bonnema G., Lindhout P. (2003). QTLs for tomato powdery mildew resistance (Oidium lycopersici) in Lycopersicon parviflorumG1.1601 co-localize with two qualitative powdery mildew resistance genes. Mol. Plant-Microbe Interact. 16, 169–176. 10.1094/MPMI.2003.16.2.169 - DOI - PubMed
-
- Bai Y., van der Hulst R., Bonnema G., Marcel T. C., Meijer-Dekens F., Niks R. E., et al. (2005). Tomato defense to Oidium neolycopersici: dominant Ol genes confer isolate-dependent resistance via a different mechanism than recessive ol-2 . Mol. Plant-Microbe Interact. 18, 354–362. 10.1094/MPMI-18-0354 - DOI - PubMed
-
- Bai Y., d. H. R., Huang C. C., Wei L., Stam P., Lindhout P. (2004). Mapping Ol-4, a gene conferring resistance to Oidium neolycopersici and originating from Lycopersicon peruvianum LA2172, requires multi-allelic, single-locus markers. Theor. Appl. Genet. 109, 1215–1223. 10.1007/s00122-004-1698-5 - DOI - PubMed
LinkOut - more resources
Full Text Sources

