Identification of core differentially methylated genes in glioma
- PMID: 31788078
- PMCID: PMC6864971
- DOI: 10.3892/ol.2019.10955
Identification of core differentially methylated genes in glioma
Abstract
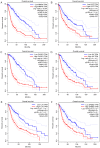
Differentially methylated genes (DMGs) serve a crucial role in the pathogenesis of glioma via the regulation of the cell cycle, proliferation, apoptosis, migration, infiltration, DNA repair and signaling pathways. This study aimed to identify aberrant DMGs and pathways by comprehensive bioinformatics analysis. The gene expression profile of GSE28094 was downloaded from the Gene Expression Omnibus (GEO) database, and the GEO2R online tool was used to find DMGs. Gene Ontology (GO) functional analysis and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of the DMGs were performed by using the Database for Annotation Visualization and Integrated Discovery. A protein-protein interaction (PPI) network was constructed with Search Tool for the Retrieval of Interacting Genes. Analysis of modules in the PPI networks was performed by Molecular Complex Detection in Cytoscape software, and four modules were performed. The hub genes with a high degree of connectivity were verified by The Cancer Genome Atlas database. A total of 349 DMGs, including 167 hypermethylation genes, were enriched in biological processes of negative and positive regulation of cell proliferation and positive regulation of transcription from RNA polymerase II promoter. Pathway analysis enrichment revealed that cancer regulated the pluripotency of stem cells and the PI3K-AKT signaling pathway, whereas 182 hypomethylated genes were enriched in biological processes of immune response, cellular response to lipopolysaccharide and peptidyl-tyrosine phosphorylation. Pathway enrichment analysis revealed cytokine-cytokine receptor interaction, type I diabetes mellitus and TNF signaling pathway. A total of 20 hub genes were identified, of which eight genes were associated with survival, including notch receptor 1 (NOTCH1), SRC proto-oncogene (also known as non-receptor tyrosine kinase, SRC), interleukin 6 (IL6), matrix metallopeptidase 9 (MMP9), interleukin 10 (IL10), caspase 3 (CASP3), erb-b2 receptor tyrosine kinase 2 (ERBB2) and epidermal growth factor (EGF). Therefore, bioinformatics analysis identified a series of core DMGs and pathways in glioma. The results of the present study may facilitate the assessment of the tumorigenicity and progression of glioma. Furthermore, the significant DMGs may provide potential methylation-based biomarkers for the precise diagnosis and targeted treatment of glioma.
Keywords: bioinformatics; biomarker; glioma; methylation.
Copyright: © Xue et al.
Figures




References
-
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
