A yeast optogenetic toolkit (yOTK) for gene expression control in Saccharomyces cerevisiae
- PMID: 31788779
- PMCID: PMC7015765
- DOI: 10.1002/bit.27234
A yeast optogenetic toolkit (yOTK) for gene expression control in Saccharomyces cerevisiae
Abstract
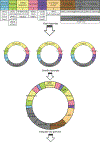
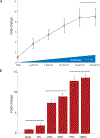
Optogenetic tools for controlling gene expression are ideal for tuning synthetic biological networks due to the exquisite spatiotemporal control available with light. Here we develop an optogenetic system for gene expression control integrated with an existing yeast toolkit allowing for rapid, modular assembly of light-controlled circuits in the important chassis organism Saccharomyces cerevisiae. We reconstitute activity of a split synthetic zinc-finger transcription factor (TF) using light-induced dimerization mediated by the proteins CRY2 and CIB1. We optimize function of this split TF and demonstrate the utility of the toolkit workflow by assembling cassettes expressing the TF activation domain and DNA-binding domain at different levels. Utilizing this TF and a synthetic promoter we demonstrate that light intensity and duty cycle can be used to modulate gene expression over the range currently available from natural yeast promoters. This study allows for rapid generation and prototyping of optogenetic circuits to control gene expression in S. cerevisiae.
Keywords: MoClo; Saccharomyces cerevisiae; light-inducible promoter; optogenetics; yeast.
© 2019 Wiley Periodicals, Inc.
Figures


References
-
- Buchholz K, & Collins J. (2013). The roots-a short history of industrial microbiology and biotechnology. Applied Microbiology and Biotechnology, 97, 3747–3762. - PubMed
-
- Carter Z, & Delneri D. (2010). New generation of loxP-mutated deletion cassettes for the genetic manipulation of yeast natural isolates. Yeast, 27, 765–775. - PubMed
-
- Chen B, Lee H, Heng Y, Chua N, Teo W, Choi W, … Chang M. (2018). Synthetic biology toolkits and applications in Saccharomyces cerevisiae. Biotechnology Advances, 36(7), 1870–1881. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

