Circulating non‑coding RNA‑biomarker potential in neoadjuvant chemotherapy of triple negative breast cancer?
- PMID: 31789396
- PMCID: PMC6910196
- DOI: 10.3892/ijo.2019.4920
Circulating non‑coding RNA‑biomarker potential in neoadjuvant chemotherapy of triple negative breast cancer?
Abstract
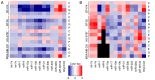
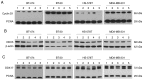
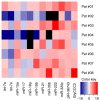
Due to the positive association between neoadjuvant chemotherapy (NACT) and the promising early response rates of patients with triple negative breast cancer (TNBC), including probabilities of pathological complete response, NACT is increasingly used in TNBC management. Liquid biopsy‑based biomarkers with the power to diagnose the early response to NACT may support established monitoring tools, which are to a certain extent imprecise and costly. Simple serum‑ or urine‑based analyses of non‑coding RNA (ncRNA) expression may allow for fast, minimally‑invasive testing and timely adjustment of the therapy regimen. The present study investigated breast cancer‑related ncRNAs [microRNA (miR)‑7, ‑9, ‑15a, ‑17, ‑18a, ‑19b, ‑21, ‑30b, ‑222 and ‑320c, PIWI‑interacting RNA‑36743 and GlyCCC2] in triple positive BT‑474 cells and three TNBC cell lines (BT‑20, HS‑578T and MDA‑MB‑231) treated with various chemotherapeutic agents using reverse transcription‑quantitative PCR. Intracellular and secreted microvesicular ncRNA expression levels were analysed using a multivariable statistical regression analysis. Chemotherapy‑driven effects were investigated by analysing cell cycle determinants at the mRNA and protein levels. Serum and urine specimens from 8 patients with TNBC were compared with 10 healthy females using two‑sample t‑tests. Samples from the patients with TNBC were compared at two time points. Chemotherapeutic treatments induced distinct changes in ncRNA expression in TNBC cell lines and the BT‑474 cell line in intra‑ and extracellular compartments. Serum and urine‑based ncRNA expression analysis was able to discriminate between patients with TNBC and controls. Time point comparisons in the urine samples of patients with TNBC revealed a general rise in the level of ncRNA. Serum data suggested a potential association between piR‑36743, miR‑17, ‑19b and ‑30b expression levels and an NACT‑driven complete clinical response. The present study highlighted the potential of ncRNAs as liquid biopsy‑based biomarkers in TNBC chemotherapy treatment. The ncRNAs tested in the present study have been previously investigated for their involvement in BC or TNBC chemotherapy responses; however, these previous studies were restricted to patient tissue or in vitro models. The data from the present study offer novel insight into ncRNA expression in liquid samples from patients with TNBC, and the study serves as an initial step in the evaluation of ncRNAs as diagnostic biomarkers in the monitoring of TNBC therapy.
Figures







References
-
- World Health Organization IAfRoC: Cancer Today. 2018 https://gco.iarc.fr/today/home. Accessed April 4, 2019.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

