Genomic Predictive Ability for Foliar Nutritive Traits in Perennial Ryegrass
- PMID: 31792009
- PMCID: PMC7003077
- DOI: 10.1534/g3.119.400880
Genomic Predictive Ability for Foliar Nutritive Traits in Perennial Ryegrass
Abstract
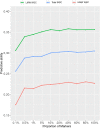
Forage nutritive value impacts animal nutrition, which underpins livestock productivity, reproduction and health. Genetic improvement for nutritive traits in perennial ryegrass has been limited, as they are typically expensive and time-consuming to measure through conventional methods. Genomic selection is appropriate for such complex and expensive traits, enabling cost-effective prediction of breeding values using genome-wide markers. The aims of the present study were to assess the potential of genomic selection for a range of nutritive traits in a multi-population training set, and to quantify contributions of family, location and family-by-location variance components to trait variation and heritability for nutritive traits. The training set consisted of a total of 517 half-sibling (half-sib) families, from five advanced breeding populations, evaluated in two distinct New Zealand grazing environments. Autumn-harvested samples were analyzed for 18 nutritive traits and maternal parents of the half-sib families were genotyped using genotyping-by-sequencing. Significant (P < 0.05) family variance was detected for all nutritive traits and genomic heritability (h2g ) was moderate to high (0.20 to 0.74). Family-by-location interactions were significant and particularly large for water soluble carbohydrate (WSC), crude fat, phosphorus (P) and crude protein. GBLUP, KGD-GBLUP and BayesCπ genomic prediction models displayed similar predictive ability, estimated by 10-fold cross validation, for all nutritive traits with values ranging from r = 0.16 to 0.45 using phenotypes from across two locations. High predictive ability was observed for the mineral traits sulfur (0.44), sodium (0.45) and magnesium (0.45) and the lowest values were observed for P (0.16), digestibility (0.22) and high molecular weight WSC (0.23). Predictive ability estimates for most nutritive traits were retained when marker number was reduced from one million to as few as 50,000. The moderate to high predictive abilities observed suggests implementation of genomic selection is feasible for most of the nutritive traits examined.
Keywords: genomic selection; heritability; nutritive traits; perennial ryegrass; water soluble carbohydrates.
Copyright © 2020 Arojju et al.
Figures

References
-
- Baert J., and Muylle H., 2016. Feeding value evaluation in grass and legume breeding and variety testing: Report of a debate, pp. 307–311 in Breeding in a World of Scarcity Springer, Berlin, Germany.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
