Exogenous Alginate Protects Staphylococcus aureus from Killing by Pseudomonas aeruginosa
- PMID: 31792010
- PMCID: PMC7099135
- DOI: 10.1128/JB.00559-19
Exogenous Alginate Protects Staphylococcus aureus from Killing by Pseudomonas aeruginosa
Abstract
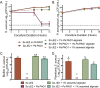
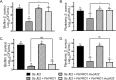
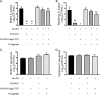
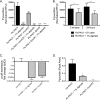
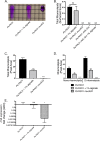
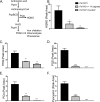
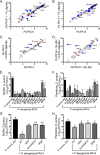
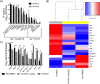
Cystic fibrosis (CF) patients chronically infected with both Pseudomonas aeruginosa and Staphylococcus aureus have worse health outcomes than patients who are monoinfected with either P. aeruginosa or S. aureus We showed previously that mucoid strains of P. aeruginosa can coexist with S. aureusin vitro due to the transcriptional downregulation of several toxic exoproducts typically produced by P. aeruginosa, including siderophores, rhamnolipids, and HQNO (2-heptyl-4-hydroxyquinoline N-oxide). Here, we demonstrate that exogenous alginate protects S. aureus from P. aeruginosa in both planktonic and biofilm coculture models under a variety of nutritional conditions. S. aureus protection in the presence of exogenous alginate is due to the transcriptional downregulation of pvdA, a gene required for the production of the iron-scavenging siderophore pyoverdine as well as the downregulation of the PQS (Pseudomonas quinolone signal) (2-heptyl-3,4-dihydroxyquinoline) quorum sensing system. The impact of exogenous alginate is independent of endogenous alginate production. We further demonstrate that coculture of mucoid P. aeruginosa with nonmucoid P. aeruginosa strains can mitigate the killing of S. aureus by the nonmucoid strain of P. aeruginosa, indicating that the mechanism that we describe here may function in vivo in the context of mixed infections. Finally, we investigated a panel of mucoid clinical isolates that retain the ability to kill S. aureus at late time points and show that each strain has a unique expression profile, indicating that mucoid isolates can overcome the S. aureus-protective effects of mucoidy in a strain-specific manner.IMPORTANCE CF patients are chronically infected by polymicrobial communities. The two dominant bacterial pathogens that infect the lungs of CF patients are P. aeruginosa and S. aureus, with ∼30% of patients coinfected by both species. Such coinfected individuals have worse outcomes than monoinfected patients, and both species persist within the same physical space. A variety of host and environmental factors have been demonstrated to promote P. aeruginosa-S. aureus coexistence, despite evidence that P. aeruginosa kills S. aureus when these organisms are cocultured in vitro Thus, a better understanding of P. aeruginosa-S. aureus interactions, particularly mechanisms by which these microorganisms are able to coexist in proximal physical space, will lead to better-informed treatments for chronic polymicrobial infections.
Keywords: HQNO; Nanostring; PQS; Pseudomonas aeruginosa; Staphylococcus aureus; alginate; cystic fibrosis; mucoid; polymicrobial; pyochelin; pyoverdine; siderophores.
Copyright © 2020 American Society for Microbiology.
Figures
Similar articles
-
Pseudomonas aeruginosa Alginate Overproduction Promotes Coexistence with Staphylococcus aureus in a Model of Cystic Fibrosis Respiratory Infection.mBio. 2017 Mar 21;8(2):e00186-17. doi: 10.1128/mBio.00186-17. mBio. 2017. PMID: 28325763 Free PMC article.
-
Pseudomonas aeruginosa Alters Staphylococcus aureus Sensitivity to Vancomycin in a Biofilm Model of Cystic Fibrosis Infection.mBio. 2017 Jul 18;8(4):e00873-17. doi: 10.1128/mBio.00873-17. mBio. 2017. PMID: 28720732 Free PMC article.
-
Genotypic and Phenotypic Diversity of Staphylococcus aureus Isolates from Cystic Fibrosis Patient Lung Infections and Their Interactions with Pseudomonas aeruginosa.mBio. 2020 Jun 23;11(3):e00735-20. doi: 10.1128/mBio.00735-20. mBio. 2020. PMID: 32576671 Free PMC article.
-
Lung infection with alginate-producing, mucoid Pseudomonas aeruginosa in cystic fibrosis.APMIS Suppl. 1992;28:1-79. APMIS Suppl. 1992. PMID: 1449848 Review.
-
Pseudomonas aeruginosa and Staphylococcus aureus communication in biofilm infections: insights through network and database construction.Crit Rev Microbiol. 2019 Sep-Nov;45(5-6):712-728. doi: 10.1080/1040841X.2019.1700209. Epub 2019 Dec 13. Crit Rev Microbiol. 2019. PMID: 31835971 Review.
Cited by
-
We have a community problem.J Bacteriol. 2024 Apr 18;206(4):e0007324. doi: 10.1128/jb.00073-24. Epub 2024 Mar 26. J Bacteriol. 2024. PMID: 38529952 Free PMC article. No abstract available.
-
Modeling reciprocal adaptation of Staphylococcus aureus and Pseudomonas aeruginosa co-isolates in artificial sputum medium.Biofilm. 2025 Apr 11;9:100279. doi: 10.1016/j.bioflm.2025.100279. eCollection 2025 Jun. Biofilm. 2025. PMID: 40290724 Free PMC article.
-
Host-microbe interactions in chronic rhinosinusitis biofilms and models for investigation.Biofilm. 2023 Sep 29;6:100160. doi: 10.1016/j.bioflm.2023.100160. eCollection 2023 Dec 15. Biofilm. 2023. PMID: 37928619 Free PMC article. Review.
-
Genomic and phenotypic characterization of Pseudomonas aeruginosa isolates from two Mexican cystic fibrosis attention centers.Microbiol Spectr. 2024 Oct 23;12(12):e0110024. doi: 10.1128/spectrum.01100-24. Online ahead of print. Microbiol Spectr. 2024. PMID: 39440985 Free PMC article.
-
Molecular Mechanisms of Staphylococcus and Pseudomonas Interactions in Cystic Fibrosis.Front Cell Infect Microbiol. 2022 Jan 6;11:824042. doi: 10.3389/fcimb.2021.824042. eCollection 2021. Front Cell Infect Microbiol. 2022. PMID: 35071057 Free PMC article. Review.
References
-
- Cystic Fibrosis Foundation. 2017. Patient registry annual data report. Cystic Fibrosis Foundation, Bethesda, MD.
-
- Rogers GB, Hart CA, Mason JR, Hughes M, Walshaw MJ, Bruce KD. 2003. Bacterial diversity in cases of lung infection in cystic fibrosis patients: 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol 41:3548–3558. doi:10.1128/jcm.41.8.3548-3558.2003. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

