DNA methylation in AgRP neurons regulates voluntary exercise behavior in mice
- PMID: 31792207
- PMCID: PMC6889160
- DOI: 10.1038/s41467-019-13339-3
DNA methylation in AgRP neurons regulates voluntary exercise behavior in mice
Abstract
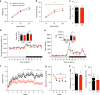
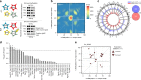
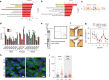
DNA methylation regulates cell type-specific gene expression. Here, in a transgenic mouse model, we show that deletion of the gene encoding DNA methyltransferase Dnmt3a in hypothalamic AgRP neurons causes a sedentary phenotype characterized by reduced voluntary exercise and increased adiposity. Whole-genome bisulfite sequencing (WGBS) and transcriptional profiling in neuronal nuclei from the arcuate nucleus of the hypothalamus (ARH) reveal differentially methylated genomic regions and reduced expression of AgRP neuron-associated genes in knockout mice. We use read-level analysis of WGBS data to infer putative ARH neural cell types affected by the knockout, and to localize promoter hypomethylation and increased expression of the growth factor Bmp7 to AgRP neurons, suggesting a role for aberrant TGF-β signaling in the development of this phenotype. Together, these data demonstrate that DNA methylation in AgRP neurons is required for their normal epigenetic development and neuron-specific gene expression profiles, and regulates voluntary exercise behavior.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

