Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors
- PMID: 31792450
- PMCID: PMC7097669
- DOI: 10.1038/s41594-019-0334-7
Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors
Abstract
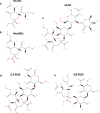
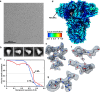
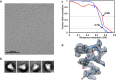
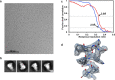
The Middle East respiratory syndrome coronavirus (MERS-CoV) causes severe and often lethal respiratory illness in humans, and no vaccines or specific treatments are available. Infections are initiated via binding of the MERS-CoV spike (S) glycoprotein to sialosides and dipeptidyl-peptidase 4 (the attachment and entry receptors, respectively). To understand MERS-CoV engagement of sialylated receptors, we determined the cryo-EM structures of S in complex with 5-N-acetyl neuraminic acid, 5-N-glycolyl neuraminic acid, sialyl-LewisX, α2,3-sialyl-N-acetyl-lactosamine and α2,6-sialyl-N-acetyl-lactosamine at 2.7-3.0 Å resolution. We show that recognition occurs via a conserved groove that is essential for MERS-CoV S-mediated attachment to sialosides and entry into human airway epithelial cells. Our data illuminate MERS-CoV S sialoside specificity and suggest that selectivity for α2,3-linked over α2,6-linked receptors results from enhanced interactions with the former class of oligosaccharides. This study provides a structural framework explaining MERS-CoV attachment to sialoside receptors and identifies a site of potential vulnerability to inhibitors of viral entry.
Conflict of interest statement
The authors declare no competing interests.
Figures
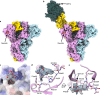


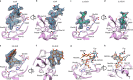




References
-
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. - PubMed
-
- Sabir JS, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

