Three-dimensional spatially resolved geometrical and functional models of human liver tissue reveal new aspects of NAFLD progression
- PMID: 31792455
- PMCID: PMC6899159
- DOI: 10.1038/s41591-019-0660-7
Three-dimensional spatially resolved geometrical and functional models of human liver tissue reveal new aspects of NAFLD progression
Abstract
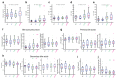
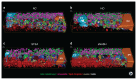
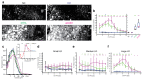
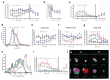
Early disease diagnosis is key to the effective treatment of diseases. Histopathological analysis of human biopsies is the gold standard to diagnose tissue alterations. However, this approach has low resolution and overlooks 3D (three-dimensional) structural changes resulting from functional alterations. Here, we applied multiphoton imaging, 3D digital reconstructions and computational simulations to generate spatially resolved geometrical and functional models of human liver tissue at different stages of non-alcoholic fatty liver disease (NAFLD). We identified a set of morphometric cellular and tissue parameters correlated with disease progression, and discover profound topological defects in the 3D bile canalicular (BC) network. Personalized biliary fluid dynamic simulations predicted an increased pericentral biliary pressure and micro-cholestasis, consistent with elevated cholestatic biomarkers in patients' sera. Our spatially resolved models of human liver tissue can contribute to high-definition medicine by identifying quantitative multiparametric cellular and tissue signatures to define disease progression and provide new insights into NAFLD pathophysiology.
Conflict of interest statement
Authors declare no competing interests
Figures
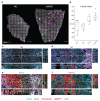

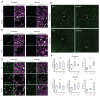
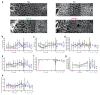

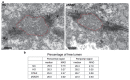


Comment in
-
High-definition medicine: modelling NAFLD in 3D.Nat Rev Gastroenterol Hepatol. 2020 Feb;17(2):66-67. doi: 10.1038/s41575-019-0256-1. Nat Rev Gastroenterol Hepatol. 2020. PMID: 31857709 No abstract available.
References
-
- Mills SE. Histology for Pathologists. Lippincott Williams & Wilkins; 2012.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

