Identification and characterization of a Masculinizer homologue in the diamondback moth, Plutella xylostella
- PMID: 31793118
- PMCID: PMC7079136
- DOI: 10.1111/imb.12628
Identification and characterization of a Masculinizer homologue in the diamondback moth, Plutella xylostella
Abstract
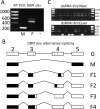
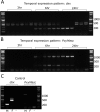
Recently, a novel sex-determination system was identified in the silkworm (Bombyx mori) in which a piwi-interacting RNA (piRNA) encoded on the female-specific W chromosome silences a Z-linked gene (Masculinizer) that would otherwise initiate male sex-determination and dosage compensation. Masculinizer provides various opportunities for developing improved genetic pest management tools. A pest lepidopteran in which a genetic pest management system has been developed, but which would benefit greatly from such improved designs, is the diamondback moth, Plutella xylostella. However, Masculinizer has not yet been identified in this species. Here, focusing on the previously described 'masculinizing' domain of B. mori Masculinizer, we identify P. xylostella Masculinizer (PxyMasc). We show that PxyMasc is Z-linked, regulates sex-specific alternative splicing of doublesex and is necessary for male survival. Similar results in B. mori suggest this survival effect is possibly through failure to initiate male dosage compensation. The highly conserved function and location of this gene between these two distantly related lepidopterans suggests a deep role for Masculinizer in the sex-determination systems of the Lepidoptera.
Keywords: Plutella xylostella; doublesex; masculinizer; diamondback moth; dosage compensation; gene drive; sex determination.
© 2019 The Authors. Insect Molecular Biology published by John Wiley & Sons Ltd on behalf of Royal Entomological Society.
Figures


Similar articles
-
Silencing RNAs expressed from W-linked PxyMasc "retrocopies" target that gene during female sex determination in Plutella xylostella.Proc Natl Acad Sci U S A. 2022 Nov 16;119(46):e2206025119. doi: 10.1073/pnas.2206025119. Epub 2022 Nov 7. Proc Natl Acad Sci U S A. 2022. PMID: 36343250 Free PMC article.
-
Masculinizer is not post-transcriptionally regulated by female-specific piRNAs during sex determination in the Asian corn borer, Ostrinia furnacalis.Insect Biochem Mol Biol. 2023 May;156:103946. doi: 10.1016/j.ibmb.2023.103946. Epub 2023 Apr 17. Insect Biochem Mol Biol. 2023. PMID: 37075905
-
A conserved role of the duplicated Masculinizer gene in sex determination of the Mediterranean flour moth, Ephestia kuehniella.PLoS Genet. 2021 Aug 2;17(8):e1009420. doi: 10.1371/journal.pgen.1009420. eCollection 2021 Aug. PLoS Genet. 2021. PMID: 34339412 Free PMC article.
-
Sex determination in moths and butterflies: Masculinizer as key player.Curr Opin Insect Sci. 2025 Aug;70:101375. doi: 10.1016/j.cois.2025.101375. Epub 2025 Apr 17. Curr Opin Insect Sci. 2025. PMID: 40252835 Review.
-
Unique sex determination system in the silkworm, Bombyx mori: current status and beyond.Proc Jpn Acad Ser B Phys Biol Sci. 2018;94(5):205-216. doi: 10.2183/pjab.94.014. Proc Jpn Acad Ser B Phys Biol Sci. 2018. PMID: 29760316 Free PMC article. Review.
Cited by
-
The Masc-PSI complex directly induces male-type doublesex splicing in silkworms.Commun Biol. 2025 Jun 14;8(1):927. doi: 10.1038/s42003-025-08350-y. Commun Biol. 2025. PMID: 40517142 Free PMC article.
-
Identification and Characterization of the Masculinizing Function of the Helicoverpa armigera Masc Gene.Int J Mol Sci. 2021 Aug 11;22(16):8650. doi: 10.3390/ijms22168650. Int J Mol Sci. 2021. PMID: 34445352 Free PMC article.
-
An Orphan Gene Enhances Male Reproductive Success in Plutella xylostella.Mol Biol Evol. 2024 Jul 3;41(7):msae142. doi: 10.1093/molbev/msae142. Mol Biol Evol. 2024. PMID: 38990889 Free PMC article.
-
Silencing RNAs expressed from W-linked PxyMasc "retrocopies" target that gene during female sex determination in Plutella xylostella.Proc Natl Acad Sci U S A. 2022 Nov 16;119(46):e2206025119. doi: 10.1073/pnas.2206025119. Epub 2022 Nov 7. Proc Natl Acad Sci U S A. 2022. PMID: 36343250 Free PMC article.
-
Developmental Shifts in the Microbiome of a Cosmopolitan Pest: Unraveling the Role of Wolbachia and Dominant Bacteria.Insects. 2024 Feb 16;15(2):132. doi: 10.3390/insects15020132. Insects. 2024. PMID: 38392551 Free PMC article.
References
-
- Alphey, N. , Bonsall, M.B. and Alphey, L. (2011) Modeling resistance to genetic control of insects. Journal of Theoretical Biology, 270, 42–55. - PubMed
-
- Backus, G.A. and Gross, K. (2016) Genetic engineering to eradicate invasive mice on islands: modelling the efficiency and ecological impacts. Ecosphere, 7, e01589.
-
- Belousova, I. , Ershov, N. , Pavlushin, S. , Ilinsky, Y. and Martemyanov, V. (2019) Molecular sexing of Lepidoptera. Journal of Insect Physiology, 114, 53–56. - PubMed
-
- Black, W.C. , Alphey, L. and James, A.A. (2011) Why RIDL is not SIT. Trends in Parasitology, 27, 362–370. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

