Dysbiosis of lower respiratory tract microbiome are associated with inflammation and microbial function variety
- PMID: 31796027
- PMCID: PMC6892239
- DOI: 10.1186/s12931-019-1246-0
Dysbiosis of lower respiratory tract microbiome are associated with inflammation and microbial function variety
Abstract
Background: Lower respiratory tract (LRT) microbiome has been reported to associate with pulmonary diseases. Unregulated inflammation is an underlying cause of variable lung diseases. The lung microbiome may play an important role in the smoking-induced inflammatory lung diseases. What's more, the function of microbiome may be more important for understanding how microbes interact with host. Our study aims to explore the effects of smoking on the lower respiratory tract microbiome, the association between variation of lower respiratory tract microbiome and inflammation and whether smoking exposure changes the function of lower respiratory tract microbime.
Methods: Forty male mice were randomly divided into smoking group and non-smoking group, and the smoking group was exposed to cigarette smoke for 2 h per day for 90 days. After experiment, the blood samples were collected to measure the concentration of interleukin-6 (IL-6) and C reactive protein (CRP) by ELISA. Lung tissue samples were used to detect the community and diversity of lower respiratory tract microbiome through 16S rRNA gene quantification and sequencing technology. ANOSIM and STAMP were performed to analyze the differences of the microbial community structure between smoking group and non-smoking group. SPSS 24.0 software was used to analyze the correlations between microbiome and inflammation mediators through scatter plots and Spearman correlation coefficient. Microbial metabolic function was predicted by PICRUSt based on the 16 s rRNA gene quantification and sequencing results. PATRIC database was searched for the potential pathogenic bacteria in lower respiratory tract.
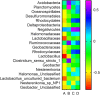
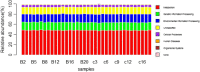
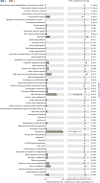
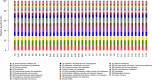
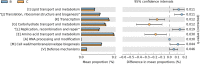
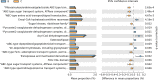
Results: Our results suggested that smoking had markedly effects on the microbiota structure of lower respiratory tract based on Bray-Curtis distance (R2 = 0.084, p = 0.005) and on unweighted uniFrac distance (R2 = 0.131, p = 0.002). Smoking mainly affected the abundance of microbiome which belong to Proteobacteria phyla and Firmicutes phyla. Moreover, our results also found that smoking increased the abundance of Acinetobacter, Bacillus and Staphylococcus, which were defined as pathogenic bacteria. Inflammatory mediators were observed to associate with certain microbiome at every level. Most of microbiome which were associated with inflammation belonged to Proteobacteria phyla or Firmicutes phyla. Moreover, we found that the decreased microbiome in smoking group, including Oceanospirillales, Desulfuromonadales, Nesterenkonia, and Lactobacillaceae, all were negatively correlated with IL-6 or CRP. Based on the level of inflammation, the abundance of microbiome differs. At genus level, Lactobacillus, Pelagibacterium, Geobacter and Zoogloea were significantly higher in smoking group with lower IL-6 concentration. The abundance of microbiome was not observed any statistical difference in subgroups with different weight. Three dominant genus, defined as pathogen, were found higher in the smoking group. The microbial functional prediction analysis revealed that ABC-type transport systems, transcription factors, amino acide transport and metabolism, arginine and proline metabolism et al. were distinctively decreased in smoking group, while the proportions of replication, recombination and repair, ribosome, DNA repair and recombination proteins were increased in smoking group (q < 0.05).
Conclusions: Members of Proteobacteria phyla and Firmicutes phyla played an important role in the microbial community composition and keeping a relatively balanced homeostasis. Microbiome dysbiosis might break the balance of immune system to drive lung inflammation. There might exist potential probiotics in lower respiratory tract, such as Lactobacillaceae. The altered function of Lower respiratory tract microbiome under smoking exposure may affect the physiological homeostasis of host.
Keywords: Function of microbiome; Inflammatory mediators; Lower respiratory tract; Microbiome; Smoking.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Wilson M. Bacteriology of humans: an ecological perspective. Malden, MA: Blackwell Publishing; 2008.
-
- Crowe SE. Helicobacter infection, chronic inflammation, and the development of malignancy. Curr Opin Gastroenterol. 2005;21(1):32–38. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

