Biased M1 receptor-positive allosteric modulators reveal role of phospholipase D in M1-dependent rodent cortical plasticity
- PMID: 31796631
- PMCID: PMC6936752
- DOI: 10.1126/scisignal.aax2057
Biased M1 receptor-positive allosteric modulators reveal role of phospholipase D in M1-dependent rodent cortical plasticity
Abstract
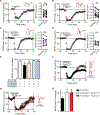
Highly selective, positive allosteric modulators (PAMs) of the M1 subtype of muscarinic acetylcholine receptor have emerged as an exciting new approach to potentially improve cognitive function in patients suffering from Alzheimer's disease and schizophrenia. Discovery programs have produced a structurally diverse range of M1 receptor PAMs with distinct pharmacological properties, including different extents of agonist activity and differences in signal bias. This includes biased M1 receptor PAMs that can potentiate coupling of the receptor to activation of phospholipase C (PLC) but not phospholipase D (PLD). However, little is known about the role of PLD in M1 receptor signaling in native systems, and it is not clear whether biased M1 PAMs display differences in modulating M1-mediated responses in native tissue. Using PLD inhibitors and PLD knockout mice, we showed that PLD was necessary for the induction of M1-dependent long-term depression (LTD) in the prefrontal cortex (PFC). Furthermore, biased M1 PAMs that did not couple to PLD not only failed to potentiate orthosteric agonist-induced LTD but also blocked M1-dependent LTD in the PFC. In contrast, biased and nonbiased M1 PAMs acted similarly in potentiating M1-dependent electrophysiological responses that were PLD independent. These findings demonstrate that PLD plays a critical role in the ability of M1 PAMs to modulate certain central nervous system (CNS) functions and that biased M1 PAMs function differently in brain regions implicated in cognition.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures
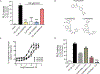



Similar articles
-
M1-positive allosteric modulators lacking agonist activity provide the optimal profile for enhancing cognition.Neuropsychopharmacology. 2018 Jul;43(8):1763-1771. doi: 10.1038/s41386-018-0033-9. Epub 2018 Mar 14. Neuropsychopharmacology. 2018. PMID: 29581537 Free PMC article.
-
Role of mGlu5 Receptors and Inhibitory Neurotransmission in M1 Dependent Muscarinic LTD in the Prefrontal Cortex: Implications in Schizophrenia.ACS Chem Neurosci. 2017 Oct 18;8(10):2254-2265. doi: 10.1021/acschemneuro.7b00167. Epub 2017 Aug 9. ACS Chem Neurosci. 2017. PMID: 28679049 Free PMC article.
-
Probing the binding site of novel selective positive allosteric modulators at the M1 muscarinic acetylcholine receptor.Biochem Pharmacol. 2018 Aug;154:243-254. doi: 10.1016/j.bcp.2018.05.009. Epub 2018 May 17. Biochem Pharmacol. 2018. PMID: 29777683 Free PMC article.
-
M1 muscarinic acetylcholine receptors: A therapeutic strategy for symptomatic and disease-modifying effects in Alzheimer's disease?Adv Pharmacol. 2020;88:277-310. doi: 10.1016/bs.apha.2019.12.003. Epub 2020 Jan 27. Adv Pharmacol. 2020. PMID: 32416870 Review.
-
Opportunities and challenges for the development of M1 muscarinic receptor positive allosteric modulators in the treatment for neurocognitive deficits.Br J Pharmacol. 2024 Jul;181(14):2114-2142. doi: 10.1111/bph.15982. Epub 2022 Dec 4. Br J Pharmacol. 2024. PMID: 36355830 Review.
Cited by
-
Fine Tuning Muscarinic Acetylcholine Receptor Signaling Through Allostery and Bias.Front Pharmacol. 2021 Jan 29;11:606656. doi: 10.3389/fphar.2020.606656. eCollection 2020. Front Pharmacol. 2021. PMID: 33584282 Free PMC article. Review.
-
Targeting Muscarinic Acetylcholine Receptors for the Treatment of Psychiatric and Neurological Disorders.Trends Pharmacol Sci. 2019 Dec;40(12):1006-1020. doi: 10.1016/j.tips.2019.10.007. Epub 2019 Nov 8. Trends Pharmacol Sci. 2019. PMID: 31711626 Free PMC article. Review.
-
Targeting the M1 muscarinic acetylcholine receptor in Alzheimer's disease.Neuronal Signal. 2022 Apr 21;6(1):NS20210004. doi: 10.1042/NS20210004. eCollection 2022 Apr. Neuronal Signal. 2022. PMID: 35571495 Free PMC article. Review.
-
Targeting muscarinic receptors to treat schizophrenia.Behav Brain Res. 2021 May 7;405:113201. doi: 10.1016/j.bbr.2021.113201. Epub 2021 Feb 26. Behav Brain Res. 2021. PMID: 33647377 Free PMC article. Review.
References
-
- Masters CL, Bateman R, Blennow K, Rowe CC, Sperling RA, Cummings JL, Alzheimer’s disease. Nat. Rev. Dis. Primers 1, 15056 (2015). - PubMed
-
- Lieberman JA, M. B, Psychotic disorders. N. Engl. J. Med 379, 270–280 (2018). - PubMed
-
- Heinrich JN, Butera JA, Carrick T, Kramer A, Kowal D, Lock T, Marquis KL, Pausch MH, Popiolek M, Sun S-C, Tseng E, Uveges AJ, Mayer SC, Pharmacological comparison of muscarinic ligands: Historical versus more recent muscarinic M1-preferring receptor agonists. Eur. J. Pharmacol 605, 53–56 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

