Estimating heritability and genetic correlations from large health datasets in the absence of genetic data
- PMID: 31796735
- PMCID: PMC6890770
- DOI: 10.1038/s41467-019-13455-0
Estimating heritability and genetic correlations from large health datasets in the absence of genetic data
Abstract
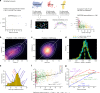
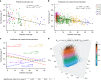
Typically, estimating genetic parameters, such as disease heritability and between-disease genetic correlations, demands large datasets containing all relevant phenotypic measures and detailed knowledge of family relationships or, alternatively, genotypic and phenotypic data for numerous unrelated individuals. Here, we suggest an alternative, efficient estimation approach through the construction of two disease metrics from large health datasets: temporal disease prevalence curves and low-dimensional disease embeddings. We present eleven thousand heritability estimates corresponding to five study types: twins, traditional family studies, health records-based family studies, single nucleotide polymorphisms, and polygenic risk scores. We also compute over six hundred thousand estimates of genetic, environmental and phenotypic correlations. Furthermore, we find that: (1) disease curve shapes cluster into five general patterns; (2) early-onset diseases tend to have lower prevalence than late-onset diseases (Spearman's ρ = 0.32, p < 10-16); and (3) the disease onset age and heritability are negatively correlated (ρ = -0.46, p < 10-16).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Cover, T. M. & Thomas, J. A. Elements of Information Theory (Wiley-Blackwell, 1991).

