Hybrid de novo whole-genome assembly and annotation of the model tapeworm Hymenolepis diminuta
- PMID: 31796747
- PMCID: PMC6890685
- DOI: 10.1038/s41597-019-0311-3
Hybrid de novo whole-genome assembly and annotation of the model tapeworm Hymenolepis diminuta
Erratum in
-
Author Correction: Hybrid de novo whole-genome assembly and annotation of the model tapeworm Hymenolepis diminuta.Sci Data. 2020 Feb 10;7(1):52. doi: 10.1038/s41597-020-0394-x. Sci Data. 2020. PMID: 32042050 Free PMC article.
Abstract
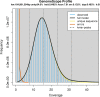
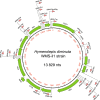
Despite the use of Hymenolepis diminuta as a model organism in experimental parasitology, a full genome description has not yet been published. Here we present a hybrid de novo genome assembly based on complementary sequencing technologies and methods. The combination of Illumina paired-end, Illumina mate-pair and Oxford Nanopore Technology reads greatly improved the assembly of the H. diminuta genome. Our results indicate that the hybrid sequencing approach is the method of choice for obtaining high-quality data. The final genome assembly is 177 Mbp with contig N50 size of 75 kbp and a scaffold N50 size of 2.3 Mbp. We obtained one of the most complete cestode genome assemblies and annotated 15,169 potential protein-coding genes. The obtained data may help explain cestode gene function and better clarify the evolution of its gene families, and thus the adaptive features evolved during millennia of co-evolution with their hosts.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Sun, T. Parasitic disorders: Pathology, diagnosis, and management. (Williams & Wilkins, 1999).
-
- Garcia, L. S. Diagnostic medical parasitology. (American Society for Microbiology Press, 2006).
-
- Skrzycki M, et al. Hymenolepis diminuta: experimental studies on the antioxidant system with short and long term infection periods in the rats. Exp. Parasitol. 2011;129:158–163. - PubMed
-
- Stradowski, M. Effects of inbreeding in Hymenolepis diminuta [Cestoda]. Acta Parasitol. 3, 146–149 (1994).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

