The C-Fern (Ceratopteris richardii) genome: insights into plant genome evolution with the first partial homosporous fern genome assembly
- PMID: 31796775
- PMCID: PMC6890710
- DOI: 10.1038/s41598-019-53968-8
The C-Fern (Ceratopteris richardii) genome: insights into plant genome evolution with the first partial homosporous fern genome assembly
Abstract
Ferns are notorious for possessing large genomes and numerous chromosomes. Despite decades of speculation, the processes underlying the expansive genomes of ferns are unclear, largely due to the absence of a sequenced homosporous fern genome. The lack of this crucial resource has not only hindered investigations of evolutionary processes responsible for the unusual genome characteristics of homosporous ferns, but also impeded synthesis of genome evolution across land plants. Here, we used the model fern species Ceratopteris richardii to address the processes (e.g., polyploidy, spread of repeat elements) by which the large genomes and high chromosome numbers typical of homosporous ferns may have evolved and have been maintained. We directly compared repeat compositions in species spanning the green plant tree of life and a diversity of genome sizes, as well as both short- and long-read-based assemblies of Ceratopteris. We found evidence consistent with a single ancient polyploidy event in the evolutionary history of Ceratopteris based on both genomic and cytogenetic data, and on repeat proportions similar to those found in large flowering plant genomes. This study provides a major stepping-stone in the understanding of land plant evolutionary genomics by providing the first homosporous fern reference genome, as well as insights into the processes underlying the formation of these massive genomes.
Conflict of interest statement
The authors declare no competing interests.
Figures
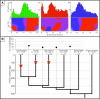
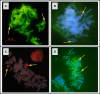
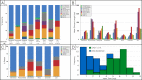
Similar articles
-
Genetic map-based analysis of genome structure in the homosporous fern Ceratopteris richardii.Genetics. 2006 Jul;173(3):1585-97. doi: 10.1534/genetics.106.055624. Epub 2006 Apr 28. Genetics. 2006. PMID: 16648591 Free PMC article.
-
Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes.BMC Evol Biol. 2009 Jun 11;9:130. doi: 10.1186/1471-2148-9-130. BMC Evol Biol. 2009. PMID: 19519899 Free PMC article.
-
An Exploration into Fern Genome Space.Genome Biol Evol. 2015 Aug 26;7(9):2533-44. doi: 10.1093/gbe/evv163. Genome Biol Evol. 2015. PMID: 26311176 Free PMC article.
-
Ever since Klekowski: testing a set of radical hypotheses revives the genetics of ferns and lycophytes.Am J Bot. 2014 Dec;101(12):2036-42. doi: 10.3732/ajb.1400317. Epub 2014 Nov 18. Am J Bot. 2014. PMID: 25480700 Review.
-
Between two fern genomes.Gigascience. 2014 Sep 25;3:15. doi: 10.1186/2047-217X-3-15. eCollection 2014. Gigascience. 2014. PMID: 25324969 Free PMC article. Review.
Cited by
-
Transcriptional analysis of Ceratopteris richardii young sporophyte reveals conservation of stem cell factors in the root apical meristem.Front Plant Sci. 2022 Aug 11;13:924660. doi: 10.3389/fpls.2022.924660. eCollection 2022. Front Plant Sci. 2022. PMID: 36035690 Free PMC article.
-
Cell Division and Meristem Dynamics in Fern Gametophytes.Plants (Basel). 2023 Jan 3;12(1):209. doi: 10.3390/plants12010209. Plants (Basel). 2023. PMID: 36616337 Free PMC article.
-
The flying spider-monkey tree fern genome provides insights into fern evolution and arborescence.Nat Plants. 2022 May;8(5):500-512. doi: 10.1038/s41477-022-01146-6. Epub 2022 May 9. Nat Plants. 2022. PMID: 35534720 Free PMC article.
-
Plant genome sequence assembly in the era of long reads: Progress, challenges and future directions.Quant Plant Biol. 2022 Mar 11;3:e5. doi: 10.1017/qpb.2021.18. eCollection 2022. Quant Plant Biol. 2022. PMID: 37077982 Free PMC article. Review.
-
Differential expression and co-localization of transcriptional factors during callus transition to differentiation for shoot organogenesis in the water fern Ceratopteris richardii.Ann Bot. 2024 Apr 10;133(3):495-507. doi: 10.1093/aob/mcae006. Ann Bot. 2024. PMID: 38206867 Free PMC article.
References
-
- Lughadha EN, et al. Counting counts: Revised estimates of numbers of accepted species of flowering plants, seed plants, vascular plants and land plants with a review of other recent estimates. Phytotaxa. 2016;272:82–88. doi: 10.11646/phytotaxa.272.1.5. - DOI
-
- Bennett, M. D. & Leitch, I. J. Plant DNA C-values database (release 6.0, Dec. 2012). WWW Doc. URL http//data.kew.org/cvalues/. [accessed 14 Oct. 2014] (2012).
-
- Pellicer J, Fay MF, Leitch IJ. The largest eukaryotic genome of them all? Bot. J. Linn. Soc. 2010;164:10–15. doi: 10.1111/j.1095-8339.2010.01072.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

