Variations in terrestrial arthropod DNA metabarcoding methods recovers robust beta diversity but variable richness and site indicators
- PMID: 31796780
- PMCID: PMC6890670
- DOI: 10.1038/s41598-019-54532-0
Variations in terrestrial arthropod DNA metabarcoding methods recovers robust beta diversity but variable richness and site indicators
Abstract
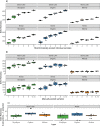
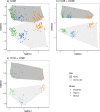
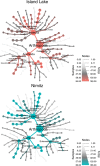
Terrestrial arthropod fauna have been suggested as a key indicator of ecological integrity in forest systems. Because phenotypic identification is expert-limited, a shift towards DNA metabarcoding could improve scalability and democratize the use of forest floor arthropods for biomonitoring applications. The objective of this study was to establish the level of field sampling and DNA extraction replication needed for arthropod biodiversity assessments from soil. Processing 15 individually collected soil samples recovered significantly higher median richness (488-614 sequence variants) than pooling the same number of samples (165-191 sequence variants) prior to DNA extraction, and we found no significant richness differences when using 1 or 3 pooled DNA extractions. Beta diversity was robust to changes in methodological regimes. Though our ability to identify taxa to species rank was limited, we were able to use arthropod COI metabarcodes from forest soil to assess richness, distinguish among sites, and recover site indicators based on unnamed exact sequence variants. Our results highlight the need to continue DNA barcoding local taxa during COI metabarcoding studies to help build reference databases. All together, these sampling considerations support the use of soil arthropod COI metabarcoding as a scalable method for biomonitoring.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Unearthing soil arthropod diversity through DNA metabarcoding.PeerJ. 2022 Feb 1;10:e12845. doi: 10.7717/peerj.12845. eCollection 2022. PeerJ. 2022. PMID: 35178296 Free PMC article.
-
Metabarcoding for biodiversity inventory blind spots: A test case using the beetle fauna of an insular cloud forest.Mol Ecol. 2023 Dec;32(23):6130-6146. doi: 10.1111/mec.16716. Epub 2022 Oct 24. Mol Ecol. 2023. PMID: 36197789
-
Establishing arthropod community composition using metabarcoding: Surprising inconsistencies between soil samples and preservative ethanol and homogenate from Malaise trap catches.Mol Ecol Resour. 2019 Nov;19(6):1516-1530. doi: 10.1111/1755-0998.13071. Epub 2019 Sep 18. Mol Ecol Resour. 2019. PMID: 31379089 Free PMC article.
-
Responses of terrestrial arthropods to air pollution: a meta-analysis.Environ Sci Pollut Res Int. 2010 Feb;17(2):297-311. doi: 10.1007/s11356-009-0138-0. Epub 2009 Mar 25. Environ Sci Pollut Res Int. 2010. PMID: 19319587 Review.
-
Plant diversity accurately predicts insect diversity in two tropical landscapes.Mol Ecol. 2016 Sep;25(17):4407-19. doi: 10.1111/mec.13770. Epub 2016 Aug 26. Mol Ecol. 2016. PMID: 27474399 Review.
Cited by
-
Comparison of traditional and DNA metabarcoding samples for monitoring tropical soil arthropods (Formicidae, Collembola and Isoptera).Sci Rep. 2022 Jun 24;12(1):10762. doi: 10.1038/s41598-022-14915-2. Sci Rep. 2022. PMID: 35750774 Free PMC article.
-
Diversity of soil faunal community as influenced by crop straw combined with different synthetic fertilizers in upland purple soil.Sci Rep. 2022 Nov 11;12(1):19306. doi: 10.1038/s41598-022-23883-6. Sci Rep. 2022. PMID: 36369353 Free PMC article.
-
Repeated subsamples during DNA extraction reveal increased diversity estimates in DNA metabarcoding of Malaise traps.Ecol Evol. 2022 Nov 27;12(11):e9502. doi: 10.1002/ece3.9502. eCollection 2022 Nov. Ecol Evol. 2022. PMID: 36447594 Free PMC article.
-
Discrepancies between prokaryotes and eukaryotes need to be considered in soil DNA-based studies.Environ Microbiol. 2022 Sep;24(9):3829-3839. doi: 10.1111/1462-2920.16019. Epub 2022 Apr 24. Environ Microbiol. 2022. PMID: 35437903 Free PMC article. Review.
-
Standardized high-throughput biomonitoring using DNA metabarcoding: Strategies for the adoption of automated liquid handlers.Environ Sci Ecotechnol. 2021 Aug 30;8:100122. doi: 10.1016/j.ese.2021.100122. eCollection 2021 Oct. Environ Sci Ecotechnol. 2021. PMID: 36156998 Free PMC article.
References
-
- Neher DA, Weicht TR, Barbercheck ME. Linking invertebrate communities to decomposition rate and nitrogen availability in pine forest soils. Applied Soil Ecology. 2012;54:14–23. doi: 10.1016/j.apsoil.2011.12.001. - DOI
-
- Maab S, Caruso T, Rillig MC. Functional role of microarthropods in soil aggregation. Pedobiologia. 2015;58:59–63. doi: 10.1016/j.pedobi.2015.03.001. - DOI
-
- Rousseau L, et al. Forest floor mesofauna communities respond to a gradient of biomass removal and soil disturbance in a boreal jack pine (Pinus banksiana) stand of northeastern Ontario (Canada) Forest Ecology and Management. 2018;407:155–165. doi: 10.1016/j.foreco.2017.08.054. - DOI
-
- Bird GA, Chatarpaul L. Effect of whole-tree and conventional forest harvest on soil microarthropods. Canadian Journal of Zoology. 1986;64:1986–1993. doi: 10.1139/z86-299. - DOI
-
- Battigelli JP, Spence JR, Langor DW, Berch SM. Short-term impact of forest soil compaction and organic matter removal on soil mesofauna density and oribatid mite diversity. Canadian Journal of Forest Research. 2004;34:1136–1149. doi: 10.1139/x03-267. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

