Human Immunodeficiency Virus (HIV)-Infected CCR6+ Rectal CD4+ T Cells and HIV Persistence On Antiretroviral Therapy
- PMID: 31796951
- PMCID: PMC7026892
- DOI: 10.1093/infdis/jiz509
Human Immunodeficiency Virus (HIV)-Infected CCR6+ Rectal CD4+ T Cells and HIV Persistence On Antiretroviral Therapy
Abstract
Background: Identifying where human immunodeficiency virus (HIV) persists in people living with HIV and receiving antiretroviral therapy is critical to develop cure strategies. We assessed the relationship of HIV persistence to expression of chemokine receptors and their chemokines in blood (n = 48) and in rectal (n = 20) and lymph node (LN; n = 8) tissue collected from people living with HIV who were receiving suppressive antiretroviral therapy.
Methods: Cell-associated integrated HIV DNA, unspliced HIV RNA, and chemokine messenger RNA were quantified by quantitative polymerase chain reaction. Chemokine receptor expression on CD4+ T cells was determined using flow cytometry.
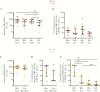
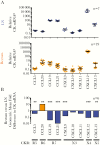
Results: Integrated HIV DNA levels in CD4+ T cells, CCR6+CXCR3+ memory CD4+ T-cell frequency, and CCL20 expression (ligand for CCR6) were highest in rectal tissue, where HIV-infected CCR6+ T cells accounted for nearly all infected cells (median, 89.7%). Conversely in LN tissue, CCR6+ T cells were infrequent, and there was a statistically significant association of cell-associated HIV DNA and RNA with CCL19, CCL21, and CXCL13 chemokines.
Conclusions: HIV-infected CCR6+ CD4+ T cells accounted for the majority of infected cells in rectal tissue. The different relationships between HIV persistence and T-cell subsets and chemokines in rectal and LN tissue suggest that different tissue-specific strategies may be required to eliminate HIV persistence and that assessment of biomarkers for HIV persistence may not be generalizable between blood and other tissues.
Keywords: CCR6; CXCR3; HIV reservoir; chemokine receptor; chemokines; latency; lymph node; persistence; rectal tissue.
© The Author(s) 2019. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures



References
-
- Chun TW, Carruth L, Finzi D, et al. . Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997; 387:183–8. - PubMed
-
- Finzi D, Hermankova M, Pierson T, et al. . Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997; 278:1295–300. - PubMed
-
- Wong JK, Hezareh M, Günthard HF, et al. . Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997; 278:1291–5. - PubMed
-
- Ganusov VV, De Boer RJ. Do most lymphocytes in humans really reside in the gut? Trends Immunol 2007; 28:514–8. - PubMed

