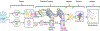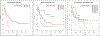Integrated Cancer Subtyping using Heterogeneous Genome-Scale Molecular Datasets
- PMID: 31797627
- PMCID: PMC6933742
Integrated Cancer Subtyping using Heterogeneous Genome-Scale Molecular Datasets
Abstract
Vast repositories of heterogeneous data from existing sources present unique opportunities. Taken individually, each of the datasets offers solutions to important domain and source-specific questions. Collectively, they represent complementary views of related data entities with an aggregate information value often well exceeding the sum of its parts. Integration of heterogeneous data is therefore paramount to i) obtain a more unified picture and comprehensive view of the relations, ii) achieve more robust results, iii) improve the accuracy and integrity, and iv) illuminate the complex interactions among data features. In this paper, we have proposed a data integration methodology to identify subtypes of cancer using multiple data types (mRNA, methylation, microRNA and somatic variants) and different data scales that come from different platforms (microarray, sequencing, etc.). The Cancer Genome Atlas (TCGA) dataset is used to build the data integration and cancer subtyping framework. The proposed data integration and disease subtyping approach accurately identifies novel subgroups of patients with significantly different survival profiles. With current availability of vast genomics, and variant data for cancer, the proposed data integration system will better differentiate cancer and patient subtypes for risk and outcome prediction and targeted treatment planning without additional cost and precious lost time.
Figures
References
-
- Saria S, Goldenberg A. “Subtyping: What It is and Its Role in Precision Medicine”, IEEE Intelligent Systems (Volume: 30, Issue: 4, July-August 2015)
-
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, et al., 2000. Molecular portraits of human breast tumours. Nature 406: 747–752. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical