Microbiome differences in disease-resistant vs. susceptible Acropora corals subjected to disease challenge assays
- PMID: 31797896
- PMCID: PMC6892807
- DOI: 10.1038/s41598-019-54855-y
Microbiome differences in disease-resistant vs. susceptible Acropora corals subjected to disease challenge assays
Abstract
In recent decades coral gardening has become increasingly popular to restore degraded reef ecosystems. However, the growth and survivorship of nursery-reared outplanted corals are highly variable. Scientists are trying to identify genotypes that show signs of disease resistance and leverage these genotypes in restoring more resilient populations. In a previous study, a field disease grafting assay was conducted on nursery-reared Acropora cervicornis and Acropora palmata to quantify relative disease susceptibility. In this study, we further evaluate this field assay by investigating putative disease-causing agents and the microbiome of corals with disease-resistant phenotypes. We conducted 16S rRNA gene high-throughput sequencing on A. cervicornis and A. palmata that were grafted (inoculated) with a diseased A. cervicornis fragment. We found that independent of health state, A. cervicornis and A. palmata had distinct alpha and beta diversity patterns from one another and distinct dominant bacteria. In addition, despite different microbiome patterns between both inoculated coral species, the genus Sphingomonadaceae was significantly found in both diseased coral species. Additionally, a core bacteria member from the order Myxococcales was found at relatively higher abundances in corals with lower rates of disease development following grafting. In all, we identified Sphingomonadaceae as a putative coral pathogen and a bacterium from the order Myxococcales associated with corals that showed disease resistant phenotypes.
Conflict of interest statement
The authors declare no competing interests.
Figures

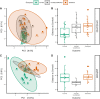

References
-
- Schopmeyer SA, et al. In Situ Coral Nurseries Serve as Genetic Repositories for Coral Reef Restoration after an Extreme Cold-Water Event. Restoration Ecology. 2012;20:696–703. doi: 10.1111/j.1526-100X.2011.00836.x. - DOI
-
- Rinkevich B. Restoration Strategies for Coral Reefs Damaged by Recreational Activities: The Use of Sexual and Asexual Recruits. Restoration Ecology. 1995;3:241–251. doi: 10.1111/j.1526-100X.1995.tb00091.x. - DOI
-
- Lirman D, et al. Propagation of the threatened staghorn coral Acropora cervicornis: methods to minimize the impacts of fragment collection and maximize production. Coral Reefs. 2010;29:729–735. doi: 10.1007/s00338-010-0621-6. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

