CD4+ T cell help creates memory CD8+ T cells with innate and help-independent recall capacities
- PMID: 31797935
- PMCID: PMC6892909
- DOI: 10.1038/s41467-019-13438-1
CD4+ T cell help creates memory CD8+ T cells with innate and help-independent recall capacities
Abstract
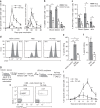
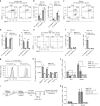
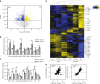
CD4+ T cell help is required for the generation of CD8+ cytotoxic T lymphocyte (CTL) memory. Here, we use genome-wide analyses to show how CD4+ T cell help delivered during priming promotes memory differentiation of CTLs. Help signals enhance IL-15-dependent maintenance of central memory T (TCM) cells. More importantly, help signals regulate the size and function of the effector memory T (TEM) cell pool. Helped TEM cells produce Granzyme B and IFNγ upon antigen-independent, innate-like recall by IL-12 and IL-18. In addition, helped memory CTLs express the effector program characteristic of helped primary CTLs upon recall with MHC class I-restricted antigens, likely due to epigenetic imprinting and sustained mRNA expression of effector genes. Our data thus indicate that during priming, CD4+ T cell help optimizes CTL memory by creating TEM cells with innate and help-independent antigen-specific recall capacities.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

