Ion Mobility Spectrometry and the Omics: Distinguishing Isomers, Molecular Classes and Contaminant Ions in Complex Samples
- PMID: 31798197
- PMCID: PMC6887105
- DOI: 10.1016/j.trac.2019.04.022
Ion Mobility Spectrometry and the Omics: Distinguishing Isomers, Molecular Classes and Contaminant Ions in Complex Samples
Abstract
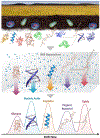
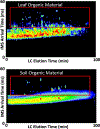
Ion mobility spectrometry (IMS) is a widely used analytical technique providing rapid gas phase separations. IMS alone is useful, but its coupling with mass spectrometry (IMS-MS) and various front-end separation techniques has greatly increased the molecular information achievable from different omic analyses. IMS-MS analyses are specifically gaining attention for improving metabolomic, lipidomic, glycomic, proteomic and exposomic analyses by increasing measurement sensitivity (e.g. S/N ratio), reducing the detection limit, and amplifying peak capacity. Numerous studies including national security-related analyses, disease screenings and environmental evaluations are illustrating that IMS-MS is able to extract information not possible with MS alone. Furthermore, IMS-MS has shown great utility in salvaging molecular information for low abundance molecules of interest when high concentration contaminant ions are present in the sample by reducing detector suppression. This review highlights how IMS-MS is currently being used in omic analyses to distinguish structurally similar molecules, isomers, molecular classes and contaminant ions.
Keywords: Exposomics; Glycomics; Ion Mobility Spectrometry; Lipidomics; Mass Spectrometry; Metabolomics; Omics; Proteomics.
Figures


References
-
- Borsdorf H, Eiceman GA. Ion Mobility Spectrometry: Principles and Applications. Applied Spectroscopy Reviews. 2006;41(4):323–75. doi: 10.1080/05704920600663469. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
