Whole Genome Analysis of Selected Human Group A Rotavirus Strains Revealed Evolution of DS-1-Like Single- and Double-Gene Reassortant Rotavirus Strains in Pakistan During 2015-2016
- PMID: 31798563
- PMCID: PMC6868104
- DOI: 10.3389/fmicb.2019.02641
Whole Genome Analysis of Selected Human Group A Rotavirus Strains Revealed Evolution of DS-1-Like Single- and Double-Gene Reassortant Rotavirus Strains in Pakistan During 2015-2016
Abstract
Acute gastroenteritis due to group A rotaviruses (RVAs) is the leading cause of infant and childhood morbidity and mortality particularly in developing countries including Pakistan. In this study we have characterized the whole genomes of five RVA strains (PAK56, PAK419, PAK585, PAK622, and PAK663) using the Illumina HiSeq platform. The strains PAK56 and PAK622 exhibited a typical Wa-like genotype constellation (G9-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1 and G3-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1, respectively), whereas PAK419, PAK585, and PAK663 exhibited distinct DS-1-like genotype constellations (G3P[4]-I2-R2-C2-M2-A2-N2-T1-E2-H2, G1P[8]-I2-R2-C2-M2-A2-N2-T2-E2-H2, and G3P[4]-I2-R2-C2-M2-A2-N2-T2-E2-H2, respectively). Despite their DS-1-like genotype constellation, strain PAK585 possessed the typical Wa-like G1P[8] genotypes, whereas both PAK419 and PAK663 possessed the G3 genotype. In addition, PAK419 also possessed the Wa-like NSP3 genotype T1, suggesting that multiple reassortments have occurred. On Phylogenetic analysis, all of the gene segments of the five strains examined in this study were genetically related to globally circulating human G1, G2, G3, G6, G8, G9, and G12 strains. Interestingly, the NSP2 gene of strain PAK419 showed closest relationship with Indian bovine strain (India/HR/B91), suggesting the occurrence of reassortment between human and bovine RVA strains. Furthermore, strains PAK419, PAK585, and PAK663 were closely related to one another in most of their gene segments, indicating that these strains might have been derived from a common ancestor. To our knowledge this is the first whole genome-based molecular characterization of human rotavirus strains in Pakistan. The results of our study will enhance our existing knowledge on the diversity and evolutionary dynamics of novel RVA strains including DS-1-like intergenogroup reassortant strains spreading in Asian countries including Pakistan, in the pre-vaccine era. Therefore, continuous surveillance is recommended to monitor the evolution, spread and genetic stability of novel reassortant rotavirus strains derived from such events.
Keywords: genotype constellation; morbidity; mortality; reassortant; surveillance.
Copyright © 2019 Sadiq, Bostan, Bokhari, Yinda and Matthijnssens.
Figures

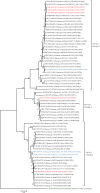


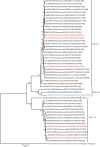







References
-
- Arana A., Montes M., Jere K. C., Alkorta M., Iturriza-Gómara M., Cilla G. (2016). Emergence and spread of G3P[8] rotaviruses possessing an equine-like VP7 and a DS-1-like genetic backbone in the Basque Country (North of Spain), 2015. Infect. Gene. Evol. 44 137–144. 10.1016/j.meegid.2016.06.048 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

