An Integrated Model of Minor Intron Emergence and Conservation
- PMID: 31798628
- PMCID: PMC6865273
- DOI: 10.3389/fgene.2019.01113
An Integrated Model of Minor Intron Emergence and Conservation
Abstract
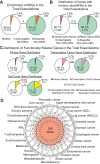
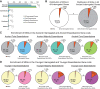
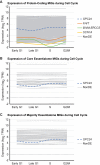
Minor introns constitute <0.5% of the introns in the human genome and have remained an enigma since their discovery. These introns are removed by a distinct splicing complex, the minor spliceosome. Both are ancient, tracing back to the last eukaryotic common ancestor (LECA), which is reflected by minor intron enrichment in specific gene families, such as the mitogen activated-protein kinase kinases, voltage-gated sodium and calcium ion channels, and E2F transcription factors. Most minor introns occur as single introns in genes with predominantly major introns. Due to this organization, minor intron-containing gene (MIG) expression requires the coordinated action of two spliceosomes, which increases the probability of missplicing. Thus, one would expect loss of minor introns via purifying selection. This has resulted in complete minor intron loss in at least nine eukaryotic lineages. However, minor introns are highly conserved in land plants and metazoans, where their importance is underscored by embryonic lethality when the minor spliceosome is inactivated. Conditional inactivation of the minor spliceosome has shown that rapidly dividing progenitor cells are highly sensitive to minor spliceosome loss. Indeed, we found that MIGs were significantly enriched in a screen for genes essential for survival in 341 cycling cell lines. Here, we propose that minor introns inserted randomly into genes in LECA or earlier and were subsequently conserved in genes crucial for cycling cell survival. We hypothesize that the essentiality of MIGs allowed minor introns to endure through the unicellularity of early eukaryotic evolution. Moreover, we identified 59 MIGs that emerged after LECA, and that many of these are essential for cycling cell survival, reinforcing our essentiality model for MIG conservation. This suggests that minor intron emergence is dynamic across eukaryotic evolution, and that minor introns should not be viewed as molecular fossils. We also posit that minor intron splicing was co-opted in multicellular evolution as a regulatory switch for en masse control of MIG expression and the biological processes they regulate. Specifically, this mode of regulation could control cell proliferation and thus body size, an idea supported by domestication syndrome, wherein MIGs are enriched in common candidate animal domestication genes.
Keywords: animal domestication; essential genes; eukaryotic evolution; human disease; minor introns; minor spliceosome; multicellularity; scaling.
Copyright © 2019 Baumgartner, Drake and Kanadia.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

