Genomic and proteomic biases inform metabolic engineering strategies for anaerobic fungi
- PMID: 31799118
- PMCID: PMC6883316
- DOI: 10.1016/j.mec.2019.e00107
Genomic and proteomic biases inform metabolic engineering strategies for anaerobic fungi
Erratum in
-
Erratum regarding previously published articles in volumes 9, 10 and 11.Metab Eng Commun. 2021 Oct 28;13:e00186. doi: 10.1016/j.mec.2021.e00186. eCollection 2021 Dec. Metab Eng Commun. 2021. PMID: 34765440 Free PMC article.
Abstract
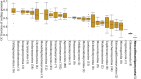
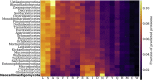
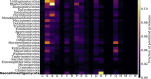
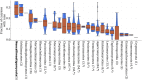
Anaerobic fungi (Neocallimastigomycota) are emerging non-model hosts for biotechnology due to their wealth of biomass-degrading enzymes, yet tools to engineer these fungi have not yet been established. Here, we show that the anaerobic gut fungi have the most GC depleted genomes among 443 sequenced organisms in the fungal kingdom, which has ramifications for heterologous expression of genes as well as for emerging CRISPR-based genome engineering approaches. Comparative genomic analyses suggest that anaerobic fungi may contain cellular machinery to aid in sexual reproduction, yet a complete mating pathway was not identified. Predicted proteomes of the anaerobic fungi also contain an unusually large fraction of proteins with homopolymeric amino acid runs consisting of five or more identical consecutive amino acids. In particular, threonine runs are especially enriched in anaerobic fungal carbohydrate active enzymes (CAZymes) and this, together with a high abundance of predicted N-glycosylation motifs, suggests that gut fungal CAZymes are heavily glycosylated, which may impact heterologous production of these biotechnologically useful enzymes. Finally, we present a codon optimization strategy to aid in the development of genetic engineering tools tailored to these early-branching anaerobic fungi.
Keywords: Amino acid distribution; Anaerobe; Codon optimization; Fungi; Genome sequencing; Neocallimastigomycota.
© 2019 The Authors.
Figures
References
-
- Albà M.M., Tompa P., Veitia R.A. Gene and Protein Evolution. KARGER; Basel: 2007. Amino acid repeats and the structure and evolution of proteins; pp. 119–130. - PubMed
-
- Arazoe T., Ogawa T., Miyoshi K., Yamato T., Ohsato S., Sakuma T., Yamamoto T., Arie T., Kuwata S. Tailor-made TALEN system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol. Bioeng. 2015;112:1335–1342. - PubMed
-
- Atasoglu C., Wallace R.J. De novo synthesis of amino acids by the ruminal anaerobic fungi, Piromyces communis and Neocallimastix frontalis. FEMS Microbiol. Lett. 2002;212:243–247. - PubMed
-
- Bach A., Calsamiglia S., Stern M.D. Nitrogen metabolism in the Rumen. J. Dairy Sci. 2005;88:E9–E21. - PubMed
-
- Beckham G.T., Bomble Y.J., Matthews J.F., Taylor C.B., Resch M.G., Yarbrough J.M., Decker S.R., Bu L., Zhao X., McCabe C., Wohlert J., Bergenstråhle M., Brady J.W., Adney W.S., Himmel M.E., Crowley M.F. The O-glycosylated linker from the Trichoderma reesei family 7 cellulase is a flexible, disordered protein. Biophys. J. 2010;99:3773–3781. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

