Medulloblastoma genomics in the modern molecular era
- PMID: 31799776
- PMCID: PMC8018047
- DOI: 10.1111/bpa.12804
Medulloblastoma genomics in the modern molecular era
Abstract
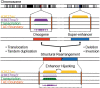
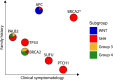
Medulloblastoma (MB) represents a spectrum of biologically and clinically distinct entities. Initially described histopathologically as a small, round blue cell tumor arising in the cerebellum, MB has emerged as a paradigm for molecular classification in cancer. Recent advances in genomic, transcriptomic and epigenomic profiling of MB have further refined molecular classification and complemented conventional histopathological diagnosis. Herein, we review the main clinical and molecular features of the four consensus subgroups of MB (WNT, SHH, Group 3 and Group 4). We also highlight hereditary predisposition syndromes associated with increased risk of MB. Finally, we explore advances in the classification of the consensus molecular groups while also presenting cutting-edge frontiers in identifying intratumoral heterogeneity and cellular origins of MB.
Keywords: genomics; medulloblastoma; molecular pathology; pediatrics.
© 2019 International Society of Neuropathology.
Figures




References
-
- Badiali M, Pession A, Basso G, Andreini L, Rigobello L, Galassi E et al (1991) N‐myc and c‐myc oncogenes amplification in medulloblastomas. Evidence of particularly aggressive behavior of a tumor with c‐myc amplification. Tumori J 77:118–121. - PubMed
-
- Bailey P, Cushing H (1925) Medulloblastoma cerebelli: a common type of midcerebellar glioma of childhood. Arch Neurol Psychiatry 14:192–224.
-
- Bigner SH, Friedman HS, Vogelstein B, Oakes WJ, Bigner DD (1990) Amplification of the c‐myc gene in human medulloblastoma cell lines and xenografts. Cancer Res 50:2347–2350. - PubMed
-
- Bunt J, Hasselt NE, Zwijnenburg DA, Hamdi M, Koster J, Versteeg R et al (2012) OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. Int J Cancer 131:E21–E32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

