Optimized Workflow for Multiplexed Phosphorylation Analysis of TMT-Labeled Peptides Using High-Field Asymmetric Waveform Ion Mobility Spectrometry
- PMID: 31799850
- PMCID: PMC6996458
- DOI: 10.1021/acs.jproteome.9b00759
Optimized Workflow for Multiplexed Phosphorylation Analysis of TMT-Labeled Peptides Using High-Field Asymmetric Waveform Ion Mobility Spectrometry
Abstract
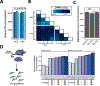
Phosphorylation is a post-translational modification with a vital role in cellular signaling. Isobaric labeling-based strategies, such as tandem mass tags (TMT), can measure the relative phosphorylation states of peptides in a multiplexed format. However, the low stoichiometry of protein phosphorylation constrains the depth of phosphopeptide analysis by mass spectrometry. As such, robust and sensitive workflows are required. Here we evaluate and optimize high-Field Asymmetric waveform Ion Mobility Spectrometry (FAIMS) coupled to Orbitrap Tribrid mass spectrometers for the analysis of TMT-labeled phosphopeptides. We determined that using FAIMS-MS3 with three compensation voltages (CV) in a single method (e.g., CV = -40/-60/-80 V) maximizes phosphopeptide coverage while minimizing inter-CV overlap. Furthermore, consecutive analyses using MSA-CID (multistage activation collision-induced dissociation) and HCD (higher-energy collisional dissociation) fragmentation at the MS2 stage increases the depth of phosphorylation analysis. The methodology and results outlined herein provide a template for tailoring optimized FAIMS-based methods.
Keywords: FAIMS; MSA; SL-TMT; SPS-MS3; TMTpro0; phosphorylation.
Figures

References
-
- Singh V; Ram M; Kumar R; Prasad R; Roy BK; Singh KK Phosphorylation: Implications in Cancer. Protein J. 2017, 36 (1), 1–6. - PubMed
-
- Manning G; Whyte DB; Martinez R; Hunter T; Sudarsanam S The protein kinase complement of the human genome. Science 2002, 298 (5600), 1912–34. - PubMed
-
- Sharma K; D’Souza RC; Tyanova S; Schaab C; Wisniewski JR; Cox J; Mann M Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014, 8 (5), 1583–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

