Dashing: fast and accurate genomic distances with HyperLogLog
- PMID: 31801633
- PMCID: PMC6892282
- DOI: 10.1186/s13059-019-1875-0
Dashing: fast and accurate genomic distances with HyperLogLog
Abstract
Dashing is a fast and accurate software tool for estimating similarities of genomes or sequencing datasets. It uses the HyperLogLog sketch together with cardinality estimation methods that are specialized for set unions and intersections. Dashing summarizes genomes more rapidly than previous MinHash-based methods while providing greater accuracy across a wide range of input sizes and sketch sizes. It can sketch and calculate pairwise distances for over 87K genomes in 6 minutes. Dashing is open source and available at https://github.com/dnbaker/dashing.
Keywords: Alignment; Genomic distance; Hyperloglog; Metagenomics; Sequencing; Sketch data structures.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

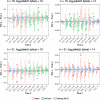
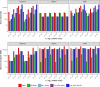

References
-
- Berlin K, Koren S, Chin CS, Drake JP, Landolin JM, Phillippy AM. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol. 2015;33(6):623–30. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

