PACSIN2 Interacts with Nonstructural Protein 5A and Regulates Hepatitis C Virus Assembly
- PMID: 31801866
- PMCID: PMC7022371
- DOI: 10.1128/JVI.01531-19
PACSIN2 Interacts with Nonstructural Protein 5A and Regulates Hepatitis C Virus Assembly
Abstract
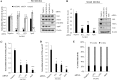
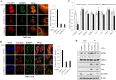
Hepatitis C virus (HCV) is a major etiologic agent of chronic liver diseases. HCV is highly dependent on cellular machinery for viral propagation. Using protein microarray analysis, we previously identified 90 cellular proteins as nonstructural 5A (NS5A) interacting partners. Of these, protein kinase C and casein kinase substrate in neurons protein 2 (PACSIN2) was selected for further study. PACSIN2 belongs to the PACSIN family, which is involved in the formation of caveolae. Protein interaction between NS5A and PACSIN2 was confirmed by pulldown assay and further verified by both coimmunoprecipitation and immunofluorescence assays. We showed that PACSIN2 interacted with domain I of NS5A and the Fer-CIP4 homology (FCH)-Bin/amphiphysin/Rvs (F-BAR) region of PACSIN2. Interestingly, NS5A specifically attenuated protein kinase C alpha (PKCα)-mediated phosphorylation of PACSIN2 at serine 313 by interrupting PACSIN2 and PKCα interaction. In fact, mutation of the serine 313 to alanine (S313A) of PACSIN2 increased protein interaction with NS5A. Silencing of PACSIN2 decreased both viral RNA and protein expression levels of HCV. Ectopic expression of the small interfering RNA (siRNA)-resistant PACSIN2 recovered the viral infectivity, suggesting that PACSIN2 was specifically required for HCV propagation. PACSIN2 was involved in viral assembly without affecting other steps of the HCV life cycle. Indeed, overexpression of PACSIN2 promoted NS5A and core protein (core) interaction. We further showed that inhibition of PKCα increased NS5A and core interaction, suggesting that phosphorylation of PACSIN2 might influence HCV assembly. Moreover, PACSIN2 was required for lipid droplet formation via modulating extracellular signal-regulated kinase 1/2 (ERK1/2) phosphorylation. Taken together, these data indicate that HCV modulates PACSIN2 via NS5A to promote virion assembly.IMPORTANCE PACSIN2 is a lipid-binding protein that triggers the tubulation of the phosphatidic acid-containing membranes. The functional involvement of PACSIN2 in the virus life cycle has not yet been demonstrated. We showed that phosphorylation of PACSIN2 displayed a negative effect on NS5A and core interaction. The most significant finding is that NS5A prevents PKCα from binding to PACSIN2. Therefore, the phosphorylation level of PACSIN2 is decreased in HCV-infected cells. We showed that HCV NS5A interrupted PKCα-mediated PACSIN2 phosphorylation at serine 313, thereby promoting NS5A-PACSIN2 interaction. We further demonstrated that PACSIN2 modulated lipid droplet formation through ERK1/2 phosphorylation. These data provide evidence that PACSIN2 is a proviral cellular factor required for viral propagation.
Keywords: NS5A; PACSIN2; hepatitis C virus; protein microarray; viral propagation.
Copyright © 2020 American Society for Microbiology.
Figures







References
-
- Bartenschlager R, Baumert TF, Bukh J, Houghton M, Lemon SM, Lindenbach BD, Lohmann V, Moradpour D, Pietschmann T, Rice CM, Thimme R, Wakita T. 2018. Critical challenges and emerging opportunities in hepatitis C virus research in an era of potent antiviral therapy: considerations for scientists and funding agencies. Virus Res 248:53–62. doi:10.1016/j.virusres.2018.02.016. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

