Cardioinformatics: the nexus of bioinformatics and precision cardiology
- PMID: 31802103
- PMCID: PMC7947182
- DOI: 10.1093/bib/bbz119
Cardioinformatics: the nexus of bioinformatics and precision cardiology
Abstract
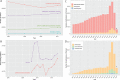
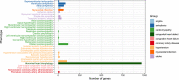
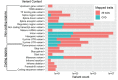
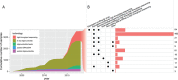
Cardiovascular disease (CVD) is the leading cause of death worldwide, causing over 17 million deaths per year, which outpaces global cancer mortality rates. Despite these sobering statistics, most bioinformatics and computational biology research and funding to date has been concentrated predominantly on cancer research, with a relatively modest footprint in CVD. In this paper, we review the existing literary landscape and critically assess the unmet need to further develop an emerging field at the multidisciplinary interface of bioinformatics and precision cardiovascular medicine, which we refer to as 'cardioinformatics'.
Keywords: bioinformatics; cardiology; cardiovascular disease; computational biology.
© The authors 2019. Published by Oxford University Press on behalf of the Institute of Mathematics and its Applications. All rights reserved.
Figures
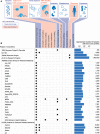
Similar articles
-
Personalized Management of Cardiovascular Disorders.Med Princ Pract. 2017;26(5):399-414. doi: 10.1159/000481403. Epub 2017 Sep 11. Med Princ Pract. 2017. PMID: 28898880 Free PMC article. Review.
-
Computational approaches to investigate the relationship between periodontitis and cardiovascular diseases for precision medicine.Hum Genomics. 2024 Oct 19;18(1):116. doi: 10.1186/s40246-024-00685-7. Hum Genomics. 2024. PMID: 39427205 Free PMC article. Review.
-
The Heart of 25 by 25: Achieving the Goal of Reducing Global and Regional Premature Deaths From Cardiovascular Diseases and Stroke: A Modeling Study From the American Heart Association and World Heart Federation.Glob Heart. 2016 Jun;11(2):251-64. doi: 10.1016/j.gheart.2016.04.002. Epub 2016 May 9. Glob Heart. 2016. PMID: 27174522
-
European Society of Cardiology: cardiovascular disease statistics 2021.Eur Heart J. 2022 Feb 22;43(8):716-799. doi: 10.1093/eurheartj/ehab892. Eur Heart J. 2022. PMID: 35016208
-
Position Paper Computational Cardiology.IEEE J Biomed Health Inform. 2019 Jan;23(1):4-11. doi: 10.1109/JBHI.2018.2877044. Epub 2018 Oct 19. IEEE J Biomed Health Inform. 2019. PMID: 30346296 Free PMC article.
Cited by
-
Multi-omics integration to identify the genetic expression and protein signature of dilated and ischemic cardiomyopathy.Front Cardiovasc Med. 2023 Feb 13;10:1115623. doi: 10.3389/fcvm.2023.1115623. eCollection 2023. Front Cardiovasc Med. 2023. PMID: 36860278 Free PMC article.
-
Beyond the Rhythm: In Silico Identification of Key Genes and Therapeutic Targets in Atrial Fibrillation.Biomedicines. 2023 Sep 25;11(10):2632. doi: 10.3390/biomedicines11102632. Biomedicines. 2023. PMID: 37893006 Free PMC article.
-
Targeting the cytoskeleton and extracellular matrix in cardiovascular disease drug discovery.Expert Opin Drug Discov. 2022 May;17(5):443-460. doi: 10.1080/17460441.2022.2047645. Epub 2022 Mar 8. Expert Opin Drug Discov. 2022. PMID: 35258387 Free PMC article. Review.
-
Novel integrated workflow allows production and in-depth quality assessment of multifactorial reprogrammed skeletal muscle cells from human stem cells.Cell Mol Life Sci. 2022 Apr 9;79(5):229. doi: 10.1007/s00018-022-04264-8. Cell Mol Life Sci. 2022. PMID: 35396689 Free PMC article.
-
HeartBioPortal2.0: new developments and updates for genetic ancestry and cardiometabolic quantitative traits in diverse human populations.Database (Oxford). 2020 Dec 31;2020:baaa115. doi: 10.1093/database/baaa115. Database (Oxford). 2020. PMID: 33382884 Free PMC article.
References
-
- Roth GA, Abate D, Abate KH, et al. Global, regional, and national age–sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018;392(10159):1736–88. doi: 10.1016/S0140-6736(18)32203-7. - DOI - DOI - PMC - PubMed
-
- Leopold JA, Loscalzo J. Emerging role of precision medicine in cardiovascular disease. Circ Res 2018;122(9):1302–15. doi: 10.1161/CIRCRESAHA.117.310782. - DOI - PMC - PubMed
-
- Heidenreich PA, Trogdon JG, Khavjou OA, et al. Forecasting the future of cardiovascular disease in the United States. Circulation 2011;123(8):933–44. doi: 10.1161/CIR.0b013e31820a55f5. - DOI - PubMed
-
- Joseph P, Leong D, McKee M, et al. Reducing the global burden of cardiovascular disease, part 1: the epidemiology and risk factors. Circ Res 2017;121(6):677–94. - PubMed
-
- Rogers MA, Aikawa E. Cardiovascular calcification: artificial intelligence and big data accelerate mechanistic discovery. Nat Rev Cardiol 2019;16(5):261–74. - PubMed

