Integrating Machine Learning With Microsimulation to Classify Hypothetical, Novel Patients for Predicting Pregabalin Treatment Response Based on Observational and Randomized Data in Patients With Painful Diabetic Peripheral Neuropathy
- PMID: 31802967
- PMCID: PMC6827520
- DOI: 10.2147/POR.S214412
Integrating Machine Learning With Microsimulation to Classify Hypothetical, Novel Patients for Predicting Pregabalin Treatment Response Based on Observational and Randomized Data in Patients With Painful Diabetic Peripheral Neuropathy
Abstract
Purpose: Variability in patient treatment responses can be a barrier to effective care. Utilization of available patient databases may improve the prediction of treatment responses. We evaluated machine learning methods to predict novel, individual patient responses to pregabalin for painful diabetic peripheral neuropathy, utilizing an agent-based modeling and simulation platform that integrates real-world observational study (OS) data and randomized clinical trial (RCT) data.
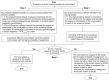
Patients and methods: The best supervised machine learning methods were selected (through literature review) and combined in a novel way for aligning patients with relevant subgroups that best enable prediction of pregabalin responses. Data were derived from a German OS of pregabalin (N=2642) and nine international RCTs (N=1320). Coarsened exact matching of OS and RCT patients was used and a hierarchical cluster analysis was implemented. We tested which machine learning methods would best align candidate patients with specific clusters that predict their pain scores over time. Cluster alignments would trigger assignments of cluster-specific time-series regressions with lagged variables as inputs in order to simulate "virtual" patients and generate 1000 trajectory variations for given novel patients.
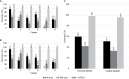
Results: Instance-based machine learning methods (k-nearest neighbor, supervised fuzzy c-means) were selected for quantitative analyses. Each method alone correctly classified 56.7% and 39.1% of patients, respectively. An "ensemble method" (combining both methods) correctly classified 98.4% and 95.9% of patients in the training and testing datasets, respectively.
Conclusion: An ensemble combination of two instance-based machine learning techniques best accommodated different data types (dichotomous, categorical, continuous) and performed better than either technique alone in assigning novel patients to subgroups for predicting treatment outcomes using microsimulation. Assignment of novel patients to a cluster of similar patients has the potential to improve prediction of patient outcomes for chronic conditions in which initial treatment response can be incorporated using microsimulation.
Clinical trial registries: www.clinicaltrials.gov: NCT00156078, NCT00159679, NCT00143156, NCT00553475.
Keywords: agent-based modeling and simulation; coarsened exact matching; hierarchical cluster analysis; machine learning; time series regressions.
© 2019 Alexander Jr et al.
Conflict of interest statement
Birol Emir, Bruce Parsons, Stephen Watt, and Ed Whalen are employees of Pfizer. Joe Alexander Jr and Marina Brodsky were employed by Pfizer at the time the study was conducted. Roger A Edwards is an employee of Health Services Consulting Corporation who was a paid consultant by Pfizer in connection with this study and development of this manuscript. Luigi Manca, Roberto Grugni, and Gianluca Bonfanti are employees of Fair Dynamics Consulting, who were paid subcontractors to Health Services Consulting Corporation in connection with this study and the development of this manuscript. The authors report no other conflicts of interest in this work.
Figures

References
Associated data
LinkOut - more resources
Full Text Sources
Medical

