Molecular evolution of peptides by yeast surface display technology
- PMID: 31803399
- PMCID: PMC6836575
- DOI: 10.1039/c9md00252a
Molecular evolution of peptides by yeast surface display technology
Abstract
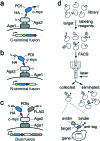
Genetically encoded peptides possess unique properties, such as a small molecular weight and ease of synthesis and modification, that make them suitable to a large variety of applications. However, despite these favorable qualities, naturally occurring peptides are often limited by intrinsic weak binding affinities, poor selectivity and low stability that ultimately restrain their final use. To overcome these limitations, a large variety of in vitro display methodologies have been developed over the past few decades to evolve genetically encoded peptide molecules with superior properties. Phage display, mRNA display, ribosome display, bacteria display, and yeast display are among the most commonly used methods to engineer peptides. While most of these in vitro methodologies have already been described in detail elsewhere, this review describes solely the yeast surface display technology and its valuable use for the evolution of a wide range of peptide formats.
This journal is © The Royal Society of Chemistry 2019.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

