Mediator MED23 regulates inflammatory responses and liver fibrosis
- PMID: 31805036
- PMCID: PMC6917294
- DOI: 10.1371/journal.pbio.3000563
Mediator MED23 regulates inflammatory responses and liver fibrosis
Abstract
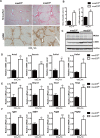
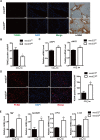
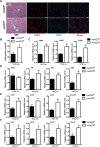
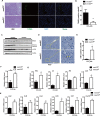
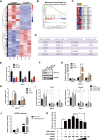
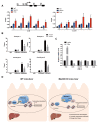
Liver fibrosis, often associated with cirrhosis and hepatocellular carcinomas, is characterized by hepatic damage, an inflammatory response, and hepatic stellate cell (HSC) activation, although the underlying mechanisms are largely unknown. Here, we show that the transcriptional Mediator complex subunit 23 (MED23) participates in the development of experimental liver fibrosis. Compared with their control littermates, mice with hepatic Med23 deletion exhibited aggravated carbon tetrachloride (CCl4)-induced liver fibrosis, with enhanced chemokine production and inflammatory infiltration as well as increased hepatocyte regeneration. Mechanistically, the orphan nuclear receptor RAR-related orphan receptor alpha (RORα) activates the expression of the liver fibrosis-related chemokines C-C motif chemokine ligand 5 (CCL5) and C-X-C motif chemokine ligand 10 (CXCL10), which is suppressed by the Mediator subunit MED23. We further found that the inhibition of Ccl5 and Cxcl10 expression by MED23 likely occurs because of G9a (also known as euchromatic histone-lysine N-methyltransferase 2 [EHMT2])-mediated H3K9 dimethylation of the target promoters. Collectively, these findings reveal hepatic MED23 as a key modulator of chemokine production and inflammatory responses and define the MED23-CCL5/CXCL10 axis as a potential target for clinical intervention in liver fibrosis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

