Human SNORA31 variations impair cortical neuron-intrinsic immunity to HSV-1 and underlie herpes simplex encephalitis
- PMID: 31806906
- PMCID: PMC7376819
- DOI: 10.1038/s41591-019-0672-3
Human SNORA31 variations impair cortical neuron-intrinsic immunity to HSV-1 and underlie herpes simplex encephalitis
Abstract
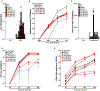
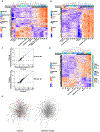
Herpes simplex virus-1 (HSV-1) encephalitis (HSE) is typically sporadic. Inborn errors of TLR3- and DBR1-mediated central nervous system cell-intrinsic immunity can account for forebrain and brainstem HSE, respectively. We report five unrelated patients with forebrain HSE, each heterozygous for one of four rare variants of SNORA31, encoding a small nucleolar RNA of the H/ACA class that are predicted to direct the isomerization of uridine residues to pseudouridine in small nuclear RNA and ribosomal RNA. We show that CRISPR/Cas9-introduced bi- and monoallelic SNORA31 deletions render human pluripotent stem cell (hPSC)-derived cortical neurons susceptible to HSV-1. Accordingly, SNORA31-mutated patient hPSC-derived cortical neurons are susceptible to HSV-1, like those from TLR3- or STAT1-deficient patients. Exogenous interferon (IFN)-β renders SNORA31- and TLR3- but not STAT1-mutated neurons resistant to HSV-1. Finally, transcriptome analysis of SNORA31-mutated neurons revealed normal responses to TLR3 and IFN-α/β stimulation but abnormal responses to HSV-1. Human SNORA31 thus controls central nervous system neuron-intrinsic immunity to HSV-1 by a distinctive mechanism.
Conflict of interest statement
Declaration of Interests
The authors have no competing interests to declare.
Figures










References
-
- Abel L, et al. Age-Dependent Mendelian Predisposition to Herpes Simplex Virus Type 1 Encephalitis in Childhood. J Pediatr 157, 623–629 (2010). - PubMed
-
- Whitley RJ Herpes simplex encephalitis: adolescents and adults. Antiviral Res 71, 141–148 (2006). - PubMed
-
- Jubelt B, Mihai C, Li TM & Veerapaneni P Rhombencephalitis / brainstem encephalitis. Current neurology and neuroscience reports 11, 543–552 (2011). - PubMed
-
- Whitley RJ Herpes Simplex Virus Infections of the Central Nervous System. Continuum 21, 1704–1713 (2015). - PubMed
Methods-only References
-
- Reed LJ & Muench H A simple method of estimating fifty percent endpoints. The American Journal of Hygiene 27, 493–497(1938).
-
- Wang Z, Liu Y, Lin W & Cui S A real-time PCR to detect and analyze virulent EMCV loads in sows and piglets. Mol Biol Rep 39, 10013–10017 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

