Identification of key candidate genes and pathways in endometrial cancer: Evidence from bioinformatics analysis
- PMID: 31807178
- PMCID: PMC6876294
- DOI: 10.3892/ol.2019.11040
Identification of key candidate genes and pathways in endometrial cancer: Evidence from bioinformatics analysis
Abstract
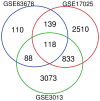
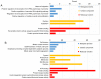
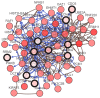
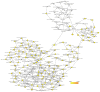
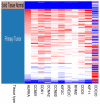
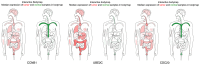
Endometrial cancer (EC) is the fourth most common cancer in women worldwide. Although researchers are exploring the biological processes of tumorigenesis and development of EC, the gene interactions and biological pathways of EC are not accurately verified. In the present study, bioinformatics methods were used to screen for key candidate genes and pathways that were associated with EC and to reveal the possible mechanisms at molecular level. Microarray datasets (GSE63678, GSE17025 and GSE3013) from the Gene Expression Omnibus database were downloaded and 118 differentially expressed genes (DEGs) were selected using a Venn diagram. Functional enrichment analyses were performed on the DEGs. A protein-protein interaction network was constructed, including the module analysis. A total of 11 hub genes were identified from the DEGs, and functional enrichment analyses were performed to clarify their possible biological processes. A total of 118 DEGs were selected from three mRNA datasets. Functional enrichment demonstrated 27 downregulated genes that were primarily involved in the positive regulation of transcription from RNA polymerase II promoter, protein binding and the nucleus. A total of 91 upregulated DEGs were mainly associated with cell division, protein binding and the nucleus. Pathway analysis indicated that the downregulated DEGs were mainly enriched in pathways associated with cancer, and the upregulated DEGs were mainly enriched in the cell cycle. The 11 hub genes were primarily enriched in the cell cycle, oocyte meiosis, progesterone-mediated oocyte maturation, the p53 signaling pathway and viral carcinogenesis. The integrated analysis showed that cyclin B1, ubiquitin conjugating enzyme E2 C and cell division cycle 20 may participate in the tumorigenesis, development and invasion of EC. In conclusion, the hub genes and pathways identified in the present study contributed to the understanding of carcinogenesis and progression of EC at the mechanistic and molecular-biological level. As candidate targets for the diagnosis and treatment of EC, these genes deserve further investigation.
Keywords: Gene Expression Omnibus database; bioinformatics analysis; differentially expressed genes; endometrial cancer.
Copyright: © Lv et al.
Figures




Similar articles
-
Identification of candidate biomarkers and pathways associated with SCLC by bioinformatics analysis.Mol Med Rep. 2018 Aug;18(2):1538-1550. doi: 10.3892/mmr.2018.9095. Epub 2018 May 29. Mol Med Rep. 2018. PMID: 29845250 Free PMC article.
-
Identification of key pathways and genes in endometrial cancer using bioinformatics analyses.Oncol Lett. 2019 Jan;17(1):897-906. doi: 10.3892/ol.2018.9667. Epub 2018 Nov 5. Oncol Lett. 2019. PMID: 30655845 Free PMC article.
-
Identification of six candidate genes for endometrial carcinoma by bioinformatics analysis.World J Surg Oncol. 2020 Jul 8;18(1):161. doi: 10.1186/s12957-020-01920-w. World J Surg Oncol. 2020. PMID: 32641130 Free PMC article.
-
Bioinformatics analysis of key differentially expressed genes in well and poorly differentiated endometrial carcinoma.Mol Med Rep. 2018 Jul;18(1):467-476. doi: 10.3892/mmr.2018.8969. Epub 2018 May 4. Mol Med Rep. 2018. PMID: 29749513
-
Identification of key genes for esophageal squamous cell carcinoma via integrated bioinformatics analysis and experimental confirmation.J Thorac Dis. 2020 Jun;12(6):3188-3199. doi: 10.21037/jtd.2020.01.33. J Thorac Dis. 2020. PMID: 32642240 Free PMC article.
Cited by
-
The Clinical Impact of Death Domain-Associated Protein and Holliday Junction Recognition Protein Expression in Cancer: Unmasking the Driving Forces of Neoplasia.Cancers (Basel). 2023 Oct 26;15(21):5165. doi: 10.3390/cancers15215165. Cancers (Basel). 2023. PMID: 37958340 Free PMC article. Review.
-
The Impact of DAXX, HJURP and CENPA Expression in Uveal Melanoma Carcinogenesis and Associations with Clinicopathological Parameters.Biomedicines. 2024 Aug 6;12(8):1772. doi: 10.3390/biomedicines12081772. Biomedicines. 2024. PMID: 39200236 Free PMC article.
-
Clinicopathologic and protein markers distinguishing the "polymerase epsilon exonuclease" from the "copy number low" subtype of endometrial cancer.J Gynecol Oncol. 2022 May;33(3):e27. doi: 10.3802/jgo.2022.33.e27. Epub 2022 Jan 17. J Gynecol Oncol. 2022. PMID: 35128857 Free PMC article.
-
Exosomal transfer of tumor-associated macrophage-derived hsa_circ_0001610 reduces radiosensitivity in endometrial cancer.Cell Death Dis. 2021 Aug 30;12(9):818. doi: 10.1038/s41419-021-04087-8. Cell Death Dis. 2021. PMID: 34462422 Free PMC article.
-
Research Progress of TXNIP as a Tumor Suppressor Gene Participating in the Metabolic Reprogramming and Oxidative Stress of Cancer Cells in Various Cancers.Front Oncol. 2020 Oct 21;10:568574. doi: 10.3389/fonc.2020.568574. eCollection 2020. Front Oncol. 2020. PMID: 33194655 Free PMC article. Review.
References
-
- Patni R. Current Concepts in Endometrial Cancer. Singapore: Springer; 2017. Endometrial carcinoma: Evolution and overview; pp. 1–9. - DOI
-
- Sheets SSF. Endometrial cancer. Surveillance, Epidemiology, and End Results Program. seer. cancer. gov. 2015
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
