Development of a Split Esterase for Protein-Protein Interaction-Dependent Small-Molecule Activation
- PMID: 31807678
- PMCID: PMC6891849
- DOI: 10.1021/acscentsci.9b00567
Development of a Split Esterase for Protein-Protein Interaction-Dependent Small-Molecule Activation
Abstract
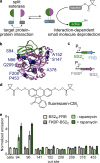
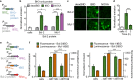
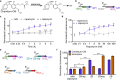
Split reporters based on fluorescent proteins and luciferases have emerged as valuable tools for measuring interactions in biological systems. Relatedly, biosensors that transduce measured input signals into outputs that influence the host system are key components of engineered gene circuits for synthetic biology applications. While small-molecule-based imaging agents are widely used in biological studies, and small-molecule-based drugs and chemical probes can target a range of biological processes, a general method for generating a target small molecule in a biological system based on a measured input signal is lacking. Here, we develop a proximity-dependent split esterase that selectively unmasks ester-protected small molecules in an interaction-dependent manner. Exploiting the versatility of an ester-protected small-molecule output, we demonstrate fluorescent, chemiluminescent, and pharmacological probe generation, each created by masking key alcohol functional groups on a target small molecule. We show that the split esterase system can be used in combination with ester-masked fluorescent or luminescent probes to measure protein-protein interactions and protein-protein interaction inhibitor engagement. We demonstrate that the esterase-based reporter system is compatible with other commonly used split reporter imaging systems for the simultaneous detection of multiple protein-protein interactions. Finally, we develop a system for selective small-molecule-dependent cell killing by unmasking a cytotoxic molecule using an inducible split esterase. Presaging utility in future synthetic biology-based therapeutic applications, we also show that the system can be used for intercellular cell killing via a bystander effect, where one activated cell unmasks a cytotoxic molecule and kills cells physically adjacent to the activated cells. Collectively, this work illustrates that the split esterase system is a valuable new addition to the split protein toolbox, with particularly exciting potential in synthetic biology applications.
Copyright © 2019 American Chemical Society.
Conflict of interest statement
The authors declare the following competing financial interest(s): A.R.L. declares a financial stake in BioLum Sciences, LLC.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

