Site-Selective C-H Halogenation Using Flavin-Dependent Halogenases Identified via Family-Wide Activity Profiling
- PMID: 31807686
- PMCID: PMC6891866
- DOI: 10.1021/acscentsci.9b00835
Site-Selective C-H Halogenation Using Flavin-Dependent Halogenases Identified via Family-Wide Activity Profiling
Abstract
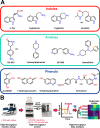
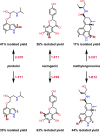
Enzymes are powerful catalysts for site-selective C-H bond functionalization. Identifying suitable enzymes for this task and for biocatalysis in general remains challenging, however, due to the fundamental difficulty of predicting catalytic activity from sequence information. In this study, family-wide activity profiling was used to obtain sequence-function information on flavin-dependent halogenases (FDHs). This broad survey provided a number of insights into FDH activity, including halide specificity and substrate preference, that were not apparent from the more focused studies reported to date. Regions of FDH sequence space that are most likely to contain enzymes suitable for halogenating small-molecule substrates were also identified. FDHs with novel substrate scope and complementary regioselectivity on large, three-dimensionally complex compounds were characterized and used for preparative-scale late-stage C-H functionalization. In many cases, these enzymes provide activities that required several rounds of directed evolution to accomplish in previous efforts, highlighting that this approach can achieve significant time savings for biocatalyst identification and provide advanced starting points for further evolution.
Copyright © 2019 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures






References
-
- Devine P. N.; Howard R. M.; Kumar R.; Thompson M. P.; Truppo M. D.; Turner N. J. Extending the Application of Biocatalysis to Meet the Challenges of Drug Development. Nat. Rev. Chem. 2018, 2, 409.10.1038/s41570-018-0055-1. - DOI
-
- Zaparucha A.; Berardinis V. d.; Vaxelaire-Vergne C.. Genome Mining for Enzyme Discovery. In Modern Biocatalysis: Advances Towards Synthetic Biological Systems; Williams G., Hall M., Eds.; The Royal Society of Chemistry: Cambridge, 2018.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

