AtFKBP53: a chimeric histone chaperone with functional nucleoplasmin and PPIase domains
- PMID: 31807785
- PMCID: PMC7026663
- DOI: 10.1093/nar/gkz1153
AtFKBP53: a chimeric histone chaperone with functional nucleoplasmin and PPIase domains
Abstract
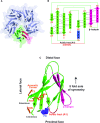
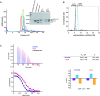
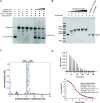
FKBP53 is one of the seven multi-domain FK506-binding proteins present in Arabidopsis thaliana, and it is known to get targeted to the nucleus. It has a conserved PPIase domain at the C-terminus and a highly charged N-terminal stretch, which has been reported to bind to histone H3 and perform the function of a histone chaperone. To better understand the molecular details of this PPIase with histone chaperoning activity, we have solved the crystal structures of its terminal domains and functionally characterized them. The C-terminal domain showed strong PPIase activity, no role in histone chaperoning and revealed a monomeric five-beta palm-like fold that wrapped over a helix, typical of an FK506-binding domain. The N-terminal domain had a pentameric nucleoplasmin-fold; making this the first report of a plant nucleoplasmin structure. Further characterization revealed the N-terminal nucleoplasmin domain to interact with H2A/H2B and H3/H4 histone oligomers, individually, as well as simultaneously, suggesting two different binding sites for H2A/H2B and H3/H4. The pentameric domain assists nucleosome assembly and forms a discrete complex with pre-formed nucleosomes; wherein two pentamers bind to a nucleosome.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Structure-function studies of a nucleoplasmin isoform from Plasmodium falciparum.J Biol Chem. 2025 Apr;301(4):108379. doi: 10.1016/j.jbc.2025.108379. Epub 2025 Mar 4. J Biol Chem. 2025. PMID: 40049416 Free PMC article.
-
The plant nucleoplasmin AtFKBP43 needs its extended arms for histone interaction.Biochim Biophys Acta Gene Regul Mech. 2022 Oct;1865(7):194872. doi: 10.1016/j.bbagrm.2022.194872. Epub 2022 Sep 2. Biochim Biophys Acta Gene Regul Mech. 2022. PMID: 36058470
-
AtFKBP53 is a histone chaperone required for repression of ribosomal RNA gene expression in Arabidopsis.Cell Res. 2010 Mar;20(3):357-66. doi: 10.1038/cr.2010.22. Epub 2010 Feb 9. Cell Res. 2010. PMID: 20142844
-
Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly.Cell Mol Life Sci. 2008 Feb;65(3):414-44. doi: 10.1007/s00018-007-7305-6. Cell Mol Life Sci. 2008. PMID: 17955179 Free PMC article. Review.
-
Histone chaperones and nucleosome assembly.Curr Opin Struct Biol. 2003 Feb;13(1):6-14. doi: 10.1016/s0959-440x(03)00002-2. Curr Opin Struct Biol. 2003. PMID: 12581654 Review.
Cited by
-
Genome-wide characterization of FK506-binding proteins, parvulins and phospho-tyrosyl phosphatase activators in wheat and their regulation by heat stress.Front Plant Sci. 2022 Dec 15;13:1053524. doi: 10.3389/fpls.2022.1053524. eCollection 2022. Front Plant Sci. 2022. PMID: 36589073 Free PMC article.
-
Structure-function studies of a nucleoplasmin isoform from Plasmodium falciparum.J Biol Chem. 2025 Apr;301(4):108379. doi: 10.1016/j.jbc.2025.108379. Epub 2025 Mar 4. J Biol Chem. 2025. PMID: 40049416 Free PMC article.
-
Genome-wide identification and expression analyses of FKBP and CYP gene family under salt and heat stress in Setaria italica L.Physiol Mol Biol Plants. 2024 Nov;30(11):1871-1887. doi: 10.1007/s12298-024-01530-w. Epub 2024 Nov 21. Physiol Mol Biol Plants. 2024. PMID: 39687704
-
Plant-specific HDT family histone deacetylases are nucleoplasmins.Plant Cell. 2022 Nov 29;34(12):4760-4777. doi: 10.1093/plcell/koac275. Plant Cell. 2022. PMID: 36069647 Free PMC article.
-
Proteomic Analysis of Generative and Vegetative Nuclei Reveals Molecular Characteristics of Pollen Cell Differentiation in Lily.Front Plant Sci. 2021 Jun 7;12:641517. doi: 10.3389/fpls.2021.641517. eCollection 2021. Front Plant Sci. 2021. PMID: 34163497 Free PMC article.
References
-
- Galat A. Peptidylprolyl cis/trans isomerases (immunophilins): biological diversity–targets–functions. Curr. Top. Med. Chem. 2003; 3:1315–1347. - PubMed
-
- Hamilton G.S., Steiner J.P.. Immunophilins: beyond immunosuppression. J. Med. Chem. 1998; 41:5119–5143. - PubMed
-
- Siekierka J.J., Wiederrecht G., Greulich H., Boulton D., Hung S.H., Cryan J., Hodges P.J., Sigal N.H.. The cytosolic-binding protein for the immunosuppressant FK-506 is both a ubiquitous and highly conserved peptidyl-prolyl cis-trans isomerase. J. Biol. Chem. 1990; 265:21011–21015. - PubMed
-
- Kang C.B., Hong Y., Dhe-Paganon S., Yoon H.S.. FKBP family proteins: immunophilins with versatile biological functions. Neurosignals. 2008; 16:318–325. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

