GH3 Auxin-Amido Synthetases Alter the Ratio of Indole-3-Acetic Acid and Phenylacetic Acid in Arabidopsis
- PMID: 31808940
- PMCID: PMC7065595
- DOI: 10.1093/pcp/pcz223
GH3 Auxin-Amido Synthetases Alter the Ratio of Indole-3-Acetic Acid and Phenylacetic Acid in Arabidopsis
Abstract
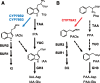
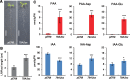
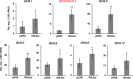
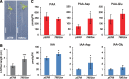
Auxin is the first discovered plant hormone and is essential for many aspects of plant growth and development. Indole-3-acetic acid (IAA) is the main auxin and plays pivotal roles in intercellular communication through polar auxin transport. Phenylacetic acid (PAA) is another natural auxin that does not show polar movement. Although a wide range of species have been shown to produce PAA, its biosynthesis, inactivation and physiological significance in plants are largely unknown. In this study, we demonstrate that overexpression of the CYP79A2 gene, which is involved in benzylglucosinolate synthesis, remarkably increased the levels of PAA and enhanced lateral root formation in Arabidopsis. This coincided with a significant reduction in the levels of IAA. The results from auxin metabolite quantification suggest that the PAA-dependent induction of GRETCHEN HAGEN 3 (GH3) genes, which encode auxin-amido synthetases, promote the inactivation of IAA. Similarly, an increase in IAA synthesis, via the indole-3-acetaldoxime pathway, significantly reduced the levels of PAA. The same adjustment of IAA and PAA levels was also observed by applying each auxin to wild-type plants. These results show that GH3 auxin-amido synthetases can alter the ratio of IAA and PAA in plant growth and development.
Keywords: Arabidopsis; Auxin; Biosynthesis; Inactivation; Indole-3-acetic acid; Phenylacetic acid.
� The Author(s) 2019. Published by Oxford University Press on behalf of Japanese Society of Plant Physiologists.
Figures
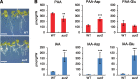
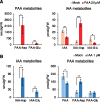
References
-
- Bejai S., Fridborg I., Ekbom B. (2012) Varied response of Spodoptera littoralis against Arabidopsis thaliana with metabolically engineered glucosinolate profiles. Plant Physiol. Biochem. 50: 72–78. - PubMed
-
- Brader G., Mikkelsen M.D., Halkier B.A., Tapio Palva E. (2006) Altering glucosinolate profiles modulates disease resistance in plants. Plant J. 46: 758–767. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

