Theory of Active Chromatin Remodeling
- PMID: 31809105
- PMCID: PMC7192239
- DOI: 10.1103/PhysRevLett.123.208102
Theory of Active Chromatin Remodeling
Abstract
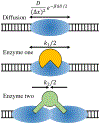
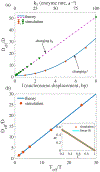
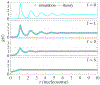
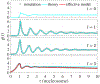
Nucleosome positioning controls the accessible regions of chromatin and plays essential roles in DNA-templated processes. ATP driven remodeling enzymes are known to be crucial for its establishment in vivo, but their nonequilibrium nature has hindered the development of a unified theoretical framework for nucleosome positioning. Using a perturbation theory, we show that the effect of these enzymes can be well approximated by effective equilibrium models with rescaled temperatures and interactions. Numerical simulations support the accuracy of the theory in predicting both kinetic and steady-state quantities, including the effective temperature and the radial distribution function, in biologically relevant regimes. The energy landscape view emerging from our study provides an intuitive understanding for the impact of remodeling enzymes in either reinforcing or overwriting intrinsic signals for nucleosome positioning, and may help improve the accuracy of computational models for its prediction in silico.
Figures

Similar articles
-
The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing.Nat Struct Mol Biol. 2006 Dec;13(12):1078-83. doi: 10.1038/nsmb1170. Epub 2006 Nov 12. Nat Struct Mol Biol. 2006. PMID: 17099699
-
Chromatin remodeling complexes: ATP-dependent machines in action.Biochem Cell Biol. 2005 Aug;83(4):405-17. doi: 10.1139/o05-115. Biochem Cell Biol. 2005. PMID: 16094444 Review.
-
ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions.Oncogene. 2001 May 28;20(24):3076-85. doi: 10.1038/sj.onc.1204332. Oncogene. 2001. PMID: 11420723 Review.
-
ATP-dependent chromatosome remodeling.Biol Chem. 2008 Apr;389(4):345-52. doi: 10.1515/BC.2008.040. Biol Chem. 2008. PMID: 18208359 Review.
-
Methods for chromatin assembly and remodeling.Methods. 2004 May;33(1):12-7. doi: 10.1016/j.ymeth.2003.10.015. Methods. 2004. PMID: 15039082
Cited by
-
Multiscale modeling of genome organization with maximum entropy optimization.J Chem Phys. 2021 Jul 7;155(1):010901. doi: 10.1063/5.0044150. J Chem Phys. 2021. PMID: 34241389 Free PMC article.
-
Interplay of chromatin organization and mechanics of the cell nucleus.Biophys J. 2024 Oct 1;123(19):3386-3396. doi: 10.1016/j.bpj.2024.08.003. Epub 2024 Aug 8. Biophys J. 2024. PMID: 39126157
-
Chromatin fiber breaks into clutches under tension and crowding.Nucleic Acids Res. 2022 Sep 23;50(17):9738-9747. doi: 10.1093/nar/gkac725. Nucleic Acids Res. 2022. PMID: 36029149 Free PMC article.
-
Compartmentalization with nuclear landmarks yields random, yet precise, genome organization.Biophys J. 2023 Apr 4;122(7):1376-1389. doi: 10.1016/j.bpj.2023.03.003. Epub 2023 Mar 5. Biophys J. 2023. PMID: 36871158 Free PMC article.
-
Molecular Determinants for the Layering and Coarsening of Biological Condensates.Aggregate (Hoboken). 2022 Dec;3(6):e306. doi: 10.1002/agt2.306. Epub 2022 Dec 10. Aggregate (Hoboken). 2022. PMID: 37065433 Free PMC article.
References
-
- Richmond RK, Sargent DF, Richmond TJ, Luger K, and Mäder AW, Crystal structure of the nucleosome core particle at 2.8 Å resolution, Nature 389, 251 (1997). - PubMed
-
- van Holde KE, Chromatin, Springer Series in Molecular Biology (Springer New York, New York, NY, 1989).
-
- Bai L and Morozov AV, Gene regulation by nucleosome positioning, Trends Genet. 26, 476 (2010). - PubMed
-
- Schiessel H, Widom J, Bruinsma RF, and Gelbart WM, Polymer Reptation and Nucleosome Repositioning, Phys. Rev. Lett 86, 4414 (2001). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
