Hic1 Defines Quiescent Mesenchymal Progenitor Subpopulations with Distinct Functions and Fates in Skeletal Muscle Regeneration
- PMID: 31809738
- PMCID: PMC6941576
- DOI: 10.1016/j.stem.2019.11.004
Hic1 Defines Quiescent Mesenchymal Progenitor Subpopulations with Distinct Functions and Fates in Skeletal Muscle Regeneration
Abstract
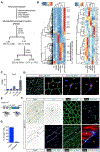
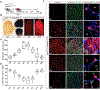
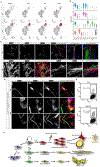
Many adult tissues contain resident stem cells, such as the Pax7+ satellite cells within skeletal muscle, that regenerate parenchymal elements following damage. Tissue-resident mesenchymal progenitors (MPs) also participate in regeneration, although their function and fate in this process are unclear. Here, we identify Hypermethylated in cancer 1 (Hic1) as a marker of MPs in skeletal muscle and further show that Hic1 deletion leads to MP hyperplasia. Single-cell RNA-seq and ATAC-seq analysis of Hic1+ MPs in skeletal muscle shows multiple subpopulations, which we further show have distinct functions and lineage potential. Hic1+ MPs orchestrate multiple aspects of skeletal muscle regeneration by providing stage-specific immunomodulation and trophic and mechanical support. During muscle regeneration, Hic1+ derivatives directly contribute to several mesenchymal compartments including Col22a1-expressing cells within the myotendinous junction. Collectively, these findings demonstrate that HIC1 regulates MP quiescence and identifies MP subpopulations with transient and enduring roles in muscle regeneration.
Keywords: lineage tracing; mesenchymal progenitors; myotendinous junction; pericytes; quiescence; scATAC-seq; scRNA-seq; skeletal muscle; tendon; tissue regeneration.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors have no competing interests to declare.
Figures




Comment in
-
Keeping Fibrotic Responses in Contractile Tissues at Bay: The Plot t(Hic1)ens.Cell Stem Cell. 2020 Feb 6;26(2):129-130. doi: 10.1016/j.stem.2019.12.010. Cell Stem Cell. 2020. PMID: 32032520
Similar articles
-
Cellular taxonomy of Hic1+ mesenchymal progenitor derivatives in the limb: from embryo to adult.Nat Commun. 2022 Aug 25;13(1):4989. doi: 10.1038/s41467-022-32695-1. Nat Commun. 2022. PMID: 36008423 Free PMC article.
-
Zfp423 Regulates Skeletal Muscle Regeneration and Proliferation.Mol Cell Biol. 2019 Apr 2;39(8):e00447-18. doi: 10.1128/MCB.00447-18. Print 2019 Apr 15. Mol Cell Biol. 2019. PMID: 30692273 Free PMC article.
-
Satellite cells, the engines of muscle repair.Nat Rev Mol Cell Biol. 2011 Dec 21;13(2):127-33. doi: 10.1038/nrm3265. Nat Rev Mol Cell Biol. 2011. PMID: 22186952 Review.
-
Hic1 identifies a specialized mesenchymal progenitor population in the embryonic limb responsible for bone superstructure formation.Cell Rep. 2023 Apr 25;42(4):112325. doi: 10.1016/j.celrep.2023.112325. Epub 2023 Mar 31. Cell Rep. 2023. PMID: 37002923
-
"Known Unknowns": Current Questions in Muscle Satellite Cell Biology.Curr Top Dev Biol. 2018;126:205-233. doi: 10.1016/bs.ctdb.2017.08.006. Epub 2017 Dec 7. Curr Top Dev Biol. 2018. PMID: 29304999 Review.
Cited by
-
Cellular taxonomy of Hic1+ mesenchymal progenitor derivatives in the limb: from embryo to adult.Nat Commun. 2022 Aug 25;13(1):4989. doi: 10.1038/s41467-022-32695-1. Nat Commun. 2022. PMID: 36008423 Free PMC article.
-
Mesenchymal progenitor cells from non-inflamed versus inflamed synovium post-ACL injury present with distinct phenotypes and cartilage regeneration capacity.Stem Cell Res Ther. 2023 Jun 25;14(1):168. doi: 10.1186/s13287-023-03396-3. Stem Cell Res Ther. 2023. PMID: 37357305 Free PMC article.
-
Assay for Transposase-Accessible Chromatin Using Sequencing of Freshly Isolated Muscle Stem Cells.Methods Mol Biol. 2023;2640:397-412. doi: 10.1007/978-1-0716-3036-5_27. Methods Mol Biol. 2023. PMID: 36995609
-
Fibro-adipogenic progenitors in physiological adipogenesis and intermuscular adipose tissue remodeling.Mol Aspects Med. 2024 Jun;97:101277. doi: 10.1016/j.mam.2024.101277. Epub 2024 May 23. Mol Aspects Med. 2024. PMID: 38788527 Free PMC article. Review.
-
Thrown for a loop: fibro-adipogenic progenitors in skeletal muscle fibrosis.Am J Physiol Cell Physiol. 2023 Oct 1;325(4):C895-C906. doi: 10.1152/ajpcell.00245.2023. Epub 2023 Aug 21. Am J Physiol Cell Physiol. 2023. PMID: 37602412 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

