ClpG Provides Increased Heat Resistance by Acting as Superior Disaggregase
- PMID: 31810333
- PMCID: PMC6995612
- DOI: 10.3390/biom9120815
ClpG Provides Increased Heat Resistance by Acting as Superior Disaggregase
Abstract
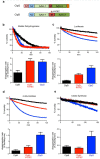
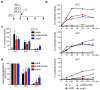
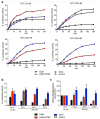
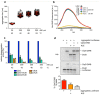
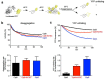
Elevation of temperature within and above the physiological limit causes the unfolding and aggregation of cellular proteins, which can ultimately lead to cell death. Bacteria are therefore equipped with Hsp100 disaggregation machines that revert the aggregation process and reactivate proteins otherwise lost by aggregation. In Gram-negative bacteria, two disaggregation systems have been described: the widespread ClpB disaggregase, which requires cooperation with an Hsp70 chaperone, and the standalone ClpG disaggregase. ClpG co-exists with ClpB in selected bacteria and provides superior heat resistance. Here, we compared the activities of both disaggregases towards diverse model substrates aggregated in vitro and in vivo at different temperatures. We show that ClpG exhibits robust activity towards all disordered aggregates, whereas ClpB acts poorly on the protein aggregates formed at very high temperatures. Extreme temperatures are expected not only to cause extended protein unfolding, but also to result in an accelerated formation of protein aggregates with potentially altered chemical and physical parameters, including increased stability. We show that ClpG exerts higher threading forces as compared to ClpB, likely enabling ClpG to process "tight" aggregates formed during severe heat stress. This defines ClpG as a more powerful disaggregase and mechanistically explains how ClpG provides increased heat resistance.
Keywords: AAA protein, Hsp100; chaperone; heat resistance; protein aggregation; protein disaggregation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

