The Biogenesis and Precise Control of RNA m6A Methylation
- PMID: 31810533
- PMCID: PMC6925345
- DOI: 10.1016/j.tig.2019.10.011
The Biogenesis and Precise Control of RNA m6A Methylation
Abstract
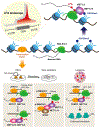
N6-Methyladenosine (m6A) is the most prevalent internal RNA modification in mRNA, and has been found to be highly conserved and hard-coded in mammals and other eukaryotic species. The importance of m6A for gene expression regulation and cell fate decisions has been well acknowledged in the past few years. However, it was only until recently that the mechanisms underlying the biogenesis and specificity of m6A modification in cells were uncovered. We review up-to-date knowledge on the biogenesis of the RNA m6A modification, including the cis-regulatory elements and trans-acting factors that determine general de novo m6A deposition and modulate cell type-specific m6A patterns, and we discuss the biological significance of such regulation.
Keywords: H3K36me3; RNA modification; biogenesis; dynamic control; epitranscriptome; m(6)A deposition.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS:
J.C. is the scientific founder of Genovel Biotech Corp.
Figures
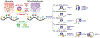
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

