Npas1+-Nkx2.1+ Neurons Are an Integral Part of the Cortico-pallido-cortical Loop
- PMID: 31811030
- PMCID: PMC6975296
- DOI: 10.1523/JNEUROSCI.1199-19.2019
Npas1+-Nkx2.1+ Neurons Are an Integral Part of the Cortico-pallido-cortical Loop
Abstract
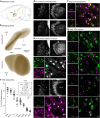
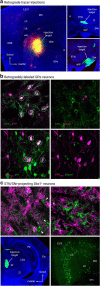
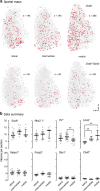
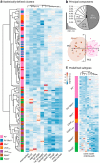
Within the basal ganglia circuit, the external globus pallidus (GPe) is critically involved in motor control. Aside from Foxp2+ neurons and ChAT+ neurons that have been established as unique neuron types, there is little consensus on the classification of GPe neurons. Properties of the remaining neuron types are poorly defined. In this study, we leverage new mouse lines, viral tools, and molecular markers to better define GPe neuron subtypes. We found that Sox6 represents a novel, defining marker for GPe neuron subtypes. Lhx6+ neurons that lack the expression of Sox6 were devoid of both parvalbumin and Npas1. This result confirms previous assertions of the existence of a unique Lhx6+ population. Neurons that arise from the Dbx1+ lineage were similarly abundant in the GPe and displayed a heterogeneous makeup. Importantly, tracing experiments revealed that Npas1+-Nkx2.1+ neurons represent the principal noncholinergic, cortically-projecting neurons. In other words, they form the pallido-cortical arm of the cortico-pallido-cortical loop. Our data further show that pyramidal-tract neurons in the cortex collateralized within the GPe, forming a closed-loop system between the two brain structures. Overall, our findings reconcile some of the discrepancies that arose from differences in techniques or the reliance on preexisting tools. Although spatial distribution and electrophysiological properties of GPe neurons reaffirm the diversification of GPe subtypes, statistical analyses strongly support the notion that these neuron subtypes can be categorized under the two principal neuron classes: PV+ neurons and Npas1+ neurons.SIGNIFICANCE STATEMENT The poor understanding of the neuronal composition in the external globus pallidus (GPe) undermines our ability to interrogate its precise behavioral and disease involvements. In this study, 12 different genetic crosses were used, hundreds of neurons were electrophysiologically characterized, and >100,000 neurons were histologically- and/or anatomically-profiled. Our current study further establishes the segregation of GPe neuron classes and illustrates the complexity of GPe neurons in adult mice. Our results support the idea that Npas1+-Nkx2.1+ neurons are a distinct GPe neuron subclass. By providing a detailed analysis of the organization of the cortico-pallidal-cortical projection, our findings establish the cellular and circuit substrates that can be important for motor function and dysfunction.
Keywords: arkypallidal neurons; basal ganglia; cellular diversity; globus pallidus; pallidocortical neurons; prototypic neurons.
Copyright © 2020 the authors.
Figures







References
-
- Abdi A, Mallet N, Mohamed FY, Sharott A, Dodson PD, Nakamura KC, Suri S, Avery SV, Larvin JT, Garas FN, Garas SN, Vinciati F, Morin S, Bezard E, Baufreton J, Magill PJ (2015) Prototypic and arkypallidal neurons in the dopamine-intact external globus pallidus. J Neurosci 35:6667–6688. 10.1523/JNEUROSCI.4662-14.2015 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 MH114829/MH/NIMH NIH HHS/United States
- P50 DA044121/DA/NIDA NIH HHS/United States
- R21 AA026022/AA/NIAAA NIH HHS/United States
- R01 MH116176/MH/NIMH NIH HHS/United States
- R01 NS069777/NS/NINDS NIH HHS/United States
- R01 MH107742/MH/NIMH NIH HHS/United States
- F32 NS098793/NS/NINDS NIH HHS/United States
- R01 NS096240/NS/NINDS NIH HHS/United States
- R01 NS088528/NS/NINDS NIH HHS/United States
- R01 NS097901/NS/NINDS NIH HHS/United States
- T32 NS041234/NS/NINDS NIH HHS/United States
- R01 MH108594/MH/NIMH NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 MH110556/MH/NIMH NIH HHS/United States
- P50 NS047085/NS/NINDS NIH HHS/United States
- ZIA MH002497/ImNIH/Intramural NIH HHS/United States
- T32 AG020506/AG/NIA NIH HHS/United States
- R01 MH112768/MH/NIMH NIH HHS/United States
- R01 MH109466/MH/NIMH NIH HHS/United States
- T32 MH067564/MH/NIMH NIH HHS/United States
- R01 NS103993/NS/NINDS NIH HHS/United States
- R56 MH114032/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
