Transcription activity contributes to the firing of non-constitutive origins in African trypanosomes helping to maintain robustness in S-phase duration
- PMID: 31811174
- PMCID: PMC6898680
- DOI: 10.1038/s41598-019-54366-w
Transcription activity contributes to the firing of non-constitutive origins in African trypanosomes helping to maintain robustness in S-phase duration
Abstract
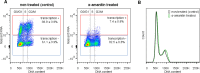
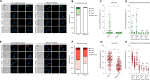
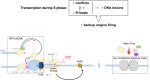
The co-synthesis of DNA and RNA potentially generates conflicts between replication and transcription, which can lead to genomic instability. In trypanosomatids, eukaryotic parasites that perform polycistronic transcription, this phenomenon and its consequences are still little studied. Here, we showed that the number of constitutive origins mapped in the Trypanosoma brucei genome is less than the minimum required to complete replication within S-phase duration. By the development of a mechanistic model of DNA replication considering replication-transcription conflicts and using immunofluorescence assays and DNA combing approaches, we demonstrated that the activation of non-constitutive (backup) origins are indispensable for replication to be completed within S-phase period. Together, our findings suggest that transcription activity during S phase generates R-loops, which contributes to the emergence of DNA lesions, leading to the firing of backup origins that help maintain robustness in S-phase duration. The usage of this increased pool of origins, contributing to the maintenance of DNA replication, seems to be of paramount importance for the survival of this parasite that affects million people around the world.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

