Computational Dissection of Membrane Transport at a Microscopic Level
- PMID: 31813734
- PMCID: PMC7024014
- DOI: 10.1016/j.tibs.2019.09.001
Computational Dissection of Membrane Transport at a Microscopic Level
Abstract
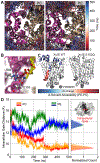
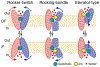
Membrane transporters are key gatekeeper proteins at cellular membranes that closely control the traffic of materials. Their function relies on structural rearrangements of varying degrees that facilitate substrate translocation across the membrane. Characterizing these functionally important molecular events at a microscopic level is key to our understanding of membrane transport, yet challenging to achieve experimentally. Recent advances in simulation technology and computing power have rendered molecular dynamics (MD) simulation a powerful biophysical tool to investigate a wide range of dynamical events spanning multiple spatial and temporal scales. Here, we review recent studies of diverse membrane transporters using computational methods, with an emphasis on highlighting the technical challenges, key lessons learned, and new opportunities to illuminate transporter structure and function.
Keywords: conformational change; drug target; free energy calculation; lipid–protein interaction; membrane transporter; molecular dynamics simulation.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Figures


References
-
- Mitchell P (1957) A general theory of membrane transport from studies of bacteria. Nature 180, 134–136 - PubMed
-
- Jardetzky O (1966) Simple allosteric model for membrane pumps. Nature 211, 969–970 - PubMed
-
- Drew D and Boudker O (2016) Shared molecular mechanisms of membrane transporters 35, 543–572 - PubMed
-
- Henderson RK et al. (2019) Coupling efficiency of secondary active transporters 58, 62–71 - PubMed
-
- Locher KP (2016) Mechanistic diversity in ATP-binding cassette (ABC) transporters 23, 487–493 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

