Galectin-3 Coordinates a Cellular System for Lysosomal Repair and Removal
- PMID: 31813797
- PMCID: PMC6997950
- DOI: 10.1016/j.devcel.2019.10.025
Galectin-3 Coordinates a Cellular System for Lysosomal Repair and Removal
Abstract
Endomembrane damage elicits homeostatic responses including ESCRT-dependent membrane repair and autophagic removal of damaged organelles. Previous studies have suggested that these systems may act separately. Here, we show that galectin-3 (Gal3), a β-galactoside-binding cytosolic lectin, unifies and coordinates ESCRT and autophagy responses to lysosomal damage. Gal3 and its capacity to recognize damage-exposed glycans were required for efficient recruitment of the ESCRT component ALIX during lysosomal damage. Both Gal3 and ALIX were required for restoration of lysosomal function. Gal3 promoted interactions between ALIX and the downstream ESCRT-III effector CHMP4 during lysosomal repair. At later time points following lysosomal injury, Gal3 controlled autophagic responses. When this failed, as in Gal3 knockout cells, lysosomal replacement program took over through TFEB. Manifestations of this staged response, which includes membrane repair, removal, and replacement, were detected in model systems of lysosomal damage inflicted by proteopathic tau and during phagosome parasitism by Mycobacterium tuberculosis.
Keywords: ESCRT; TFEB; autophagy; endosome; galectins; lysosome; membrane damage homeostasis.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures

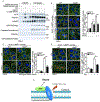
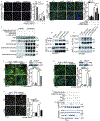

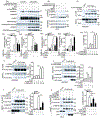

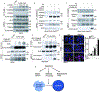
References
-
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. (2012). Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol 14, 677–685. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

