SPP1 functions as an enhancer of cell growth in hepatocellular carcinoma targeted by miR-181c
- PMID: 31814897
- PMCID: PMC6895505
SPP1 functions as an enhancer of cell growth in hepatocellular carcinoma targeted by miR-181c
Abstract
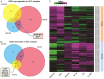
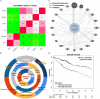
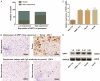
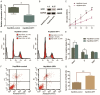
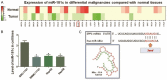
Patients diagnosed with hepatocellular carcinoma (HCC) suffered a high risk of recurrence and poor prognosis. Identification of differentially expressed genes (DEGs) in HCC provides potential biomarkers for evaluating prognosis and specific therapeutic treatments. In this study, DEGs over-expressed in HCC specimens with a fold change over 2.0 were collected through integrative bioinformatics analysis from GEO datasets. Gene ontology and KEGG pathway enrichment were conducted by applying DAVID database. We noticed Secreted phosphoprotein 1 (SPP1) as one of the signature genes up-regulated in HCC tissues with a close relation to the tumor process. Eighty-seven paired HCC specimens from our medical center were explored to verify the aberrant expression of SPP1 by IHC and qRT-PCR assay. Depletion of SPP1 in HCC Hep3B cells was established. The cell proliferation was impaired in SPP1 depleted cells, along with a resistance of cell apoptosis by down-regulating SPP1. Intriguingly, we further validated a direct interaction between miR-181c and SPP1, which indicated a post-transcriptional regulation mechanism of SPP1 in HCC. Thus, our results suggest that SPP1 may function as an enhancer of HCC growth targeted by miR-181c, and probably provide us an innovational target for HCC diagnose and therapeutic treatment.
Keywords: Hepatocellular carcinoma; cell growth; miR-181c; secreted phosphoprotein 1.
AJTR Copyright © 2019.
Conflict of interest statement
None.
Figures

Similar articles
-
Integrative bioinformatics analysis identifies ROBO1 as a potential therapeutic target modified by miR-218 in hepatocellular carcinoma.Oncotarget. 2017 May 23;8(37):61327-61337. doi: 10.18632/oncotarget.18099. eCollection 2017 Sep 22. Oncotarget. 2017. PMID: 28977866 Free PMC article.
-
Down-regulation of miR-26a-5p in hepatocellular carcinoma: A qRT-PCR and bioinformatics study.Pathol Res Pract. 2017 Dec;213(12):1494-1509. doi: 10.1016/j.prp.2017.10.001. Epub 2017 Oct 10. Pathol Res Pract. 2017. PMID: 29113686
-
MicroRNA‑181c suppresses growth and metastasis of hepatocellular carcinoma by modulating NCAPG.Cancer Manag Res. 2019 Apr 23;11:3455-3467. doi: 10.2147/CMAR.S197716. eCollection 2019. Cancer Manag Res. 2019. PMID: 31114379 Free PMC article.
-
SPP1, analyzed by bioinformatics methods, promotes the metastasis in colorectal cancer by activating EMT pathway.Biomed Pharmacother. 2017 Jul;91:1167-1177. doi: 10.1016/j.biopha.2017.05.056. Epub 2017 May 17. Biomed Pharmacother. 2017. PMID: 28531945
-
Identification and interaction analysis of key genes and microRNAs in hepatocellular carcinoma by bioinformatics analysis.World J Surg Oncol. 2017 Mar 16;15(1):63. doi: 10.1186/s12957-017-1127-2. World J Surg Oncol. 2017. PMID: 28302149 Free PMC article.
Cited by
-
Evaluation of aliphatic acid metabolism in bladder cancer with the goal of guiding therapeutic treatment.Front Oncol. 2022 Aug 18;12:930038. doi: 10.3389/fonc.2022.930038. eCollection 2022. Front Oncol. 2022. PMID: 36059672 Free PMC article.
-
Identification of biomarkers for hepatocellular carcinoma based on single cell sequencing and machine learning algorithms.Front Genet. 2022 Oct 24;13:873218. doi: 10.3389/fgene.2022.873218. eCollection 2022. Front Genet. 2022. PMID: 36353113 Free PMC article.
-
In Silico and In Vivo Evaluation of microRNA-181c-5p's Role in Hepatocellular Carcinoma.Genes (Basel). 2022 Dec 12;13(12):2343. doi: 10.3390/genes13122343. Genes (Basel). 2022. PMID: 36553610 Free PMC article.
-
Comprehensive genomic signature of pyroptosis-related genes and relevant characterization in hepatocellular carcinoma.PeerJ. 2023 Jan 12;11:e14691. doi: 10.7717/peerj.14691. eCollection 2023. PeerJ. 2023. PMID: 36650832 Free PMC article.
-
A Novel Expression Signature from the Perspective of Mesenchymal-Epithelial Transition for Hepatocellular Carcinoma with Regard to Prognosis, Clinicopathological Features, Immune Cell Infiltration, Chemotherapeutic Efficacy, and Immunosuppressive Molecules.J Oncol. 2021 Jul 28;2021:5033416. doi: 10.1155/2021/5033416. eCollection 2021. J Oncol. 2021. PMID: 34367283 Free PMC article.
References
-
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. - PubMed
-
- Reig M, Mariño Z, Perelló C, Iñarrairaegui M, Ribeiro A, Lens S, Díaz A, Vilana R, Darnell A, Varela M, Sangro B, Calleja JL, Forns X, Bruix J. Unexpected high rate of early tumor recurrence in patients with HCV-related HCC undergoing interferon-free therapy. J Hepatol. 2016;65:719–726. - PubMed
-
- Nishibatake Kinoshita M, Minami T, Tateishi R, Wake T, Nakagomi R, Fujiwara N, Sato M, Uchino K, Enooku K, Nakagawa H, Asaoka Y, Shiina S, Koike K. Impact of direct-acting antivirals on early recurrence of HCV-related HCC: comparison with interferon-based therapy. J Hepatol. 2019;70:78–86. - PubMed
-
- Shen S, Kong J, Qiu Y, Yang X, Wang W, Yan L. Identification of core genes and outcomes in hepatocellular carcinoma by bioinformatics analysis. J Cell Biochem. 2019;120:10069–10081. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
