NCAM regulates temporal specification of neural progenitor cells via profilin2 during corticogenesis
- PMID: 31816056
- PMCID: PMC7039204
- DOI: 10.1083/jcb.201902164
NCAM regulates temporal specification of neural progenitor cells via profilin2 during corticogenesis
Abstract
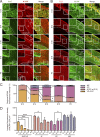
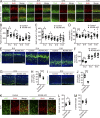
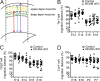
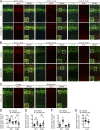
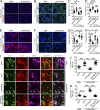
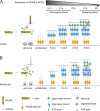
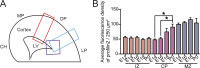
The development of cerebral cortex requires spatially and temporally orchestrated proliferation, migration, and differentiation of neural progenitor cells (NPCs). The molecular mechanisms underlying cortical development are, however, not fully understood. The neural cell adhesion molecule (NCAM) has been suggested to play a role in corticogenesis. Here we show that NCAM is dynamically expressed in the developing cortex. NCAM expression in NPCs is highest in the neurogenic period and declines during the gliogenic period. In mice bearing an NPC-specific NCAM deletion, proliferation of NPCs is reduced, and production of cortical neurons is delayed, while formation of cortical glia is advanced. Mechanistically, NCAM enhances actin polymerization in NPCs by interacting with actin-associated protein profilin2. NCAM-dependent regulation of NPCs is blocked by mutations in the profilin2 binding site. Thus, NCAM plays an essential role in NPC proliferation and fate decision during cortical development by regulating profilin2-dependent actin polymerization.
© 2019 Huang et al.
Figures







Similar articles
-
Functional cross-talk between the cellular prion protein and the neural cell adhesion molecule is critical for neuronal differentiation of neural stem/precursor cells.Stem Cells. 2014 Jun;32(6):1674-87. doi: 10.1002/stem.1663. Stem Cells. 2014. PMID: 24497115
-
Tuba8 Drives Differentiation of Cortical Radial Glia into Apical Intermediate Progenitors by Tuning Modifications of Tubulin C Termini.Dev Cell. 2020 Feb 24;52(4):477-491.e8. doi: 10.1016/j.devcel.2020.01.036. Dev Cell. 2020. PMID: 32097653 Free PMC article.
-
Bmp7 regulates the survival, proliferation, and neurogenic properties of neural progenitor cells during corticogenesis in the mouse.PLoS One. 2012;7(3):e34088. doi: 10.1371/journal.pone.0034088. Epub 2012 Mar 26. PLoS One. 2012. PMID: 22461901 Free PMC article.
-
Epigenetics and epitranscriptomics in temporal patterning of cortical neural progenitor competence.J Cell Biol. 2018 Jun 4;217(6):1901-1914. doi: 10.1083/jcb.201802117. Epub 2018 Apr 17. J Cell Biol. 2018. PMID: 29666150 Free PMC article. Review.
-
How neural stem cells contribute to neocortex development.Biochem Soc Trans. 2021 Nov 1;49(5):1997-2006. doi: 10.1042/BST20200923. Biochem Soc Trans. 2021. PMID: 34397081 Free PMC article. Review.
Cited by
-
Profilin 2 isoform expression is associated with lung metastasis of colorectal cancer according to a comprehensive gene expression study using a mouse model.Oncol Lett. 2024 Jun 17;28(2):381. doi: 10.3892/ol.2024.14514. eCollection 2024 Aug. Oncol Lett. 2024. PMID: 38939626 Free PMC article.
-
Mild hypothermia enhances regenerative gene expression in late-stage neural precursors.Med Int (Lond). 2025 Jul 16;5(5):53. doi: 10.3892/mi.2025.252. eCollection 2025 Sep-Oct. Med Int (Lond). 2025. PMID: 40747152 Free PMC article.
-
Neural glycomics: the sweet side of nervous system functions.Cell Mol Life Sci. 2021 Jan;78(1):93-116. doi: 10.1007/s00018-020-03578-9. Epub 2020 Jul 1. Cell Mol Life Sci. 2021. PMID: 32613283 Free PMC article. Review.
-
NCAM mimetic peptide P2 synergizes with bone marrow mesenchymal stem cells in promoting functional recovery after stroke.J Cereb Blood Flow Metab. 2024 Jul;44(7):1128-1144. doi: 10.1177/0271678X241226482. Epub 2024 Jan 17. J Cereb Blood Flow Metab. 2024. PMID: 38230663 Free PMC article.
-
Non-Invasive Prenatal Screening for Down Syndrome: A Review of Mass-Spectrometry-Based Approaches.Life (Basel). 2025 Apr 24;15(5):695. doi: 10.3390/life15050695. Life (Basel). 2025. PMID: 40430124 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

