Single-cell analysis of RORα tracer mouse lung reveals ILC progenitors and effector ILC2 subsets
- PMID: 31816636
- PMCID: PMC7062532
- DOI: 10.1084/jem.20182293
Single-cell analysis of RORα tracer mouse lung reveals ILC progenitors and effector ILC2 subsets
Abstract
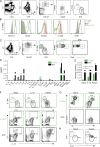
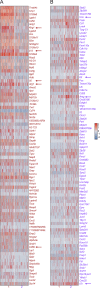
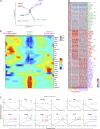
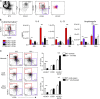
Lung group 2 innate lymphoid cells (ILC2s) drive allergic inflammation and promote tissue repair. ILC2 development is dependent on the transcription factor retinoic acid receptor-related orphan receptor (RORα), which is also expressed in common ILC progenitors. To elucidate the developmental pathways of lung ILC2s, we generated RORα lineage tracer mice and performed single-cell RNA sequencing, flow cytometry, and functional analyses. In adult mouse lungs, we found an IL-18Rα+ST2- population different from conventional IL-18Rα-ST2+ ILC2s. The former was GATA-3intTcf7EGFP+Kit+, produced few cytokines, and differentiated into multiple ILC lineages in vivo and in vitro. In neonatal mouse lungs, three ILC populations were identified, namely an ILC progenitor population similar to that in adult lungs and two distinct effector ILC2 subsets that differentially produced type 2 cytokines and amphiregulin. Lung ILC progenitors might actively contribute to ILC-poiesis in neonatal and inflamed adult lungs. In addition, neonatal lung ILC2s include distinct proinflammatory and tissue-repairing subsets.
© 2019 Ghaedi et al.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

